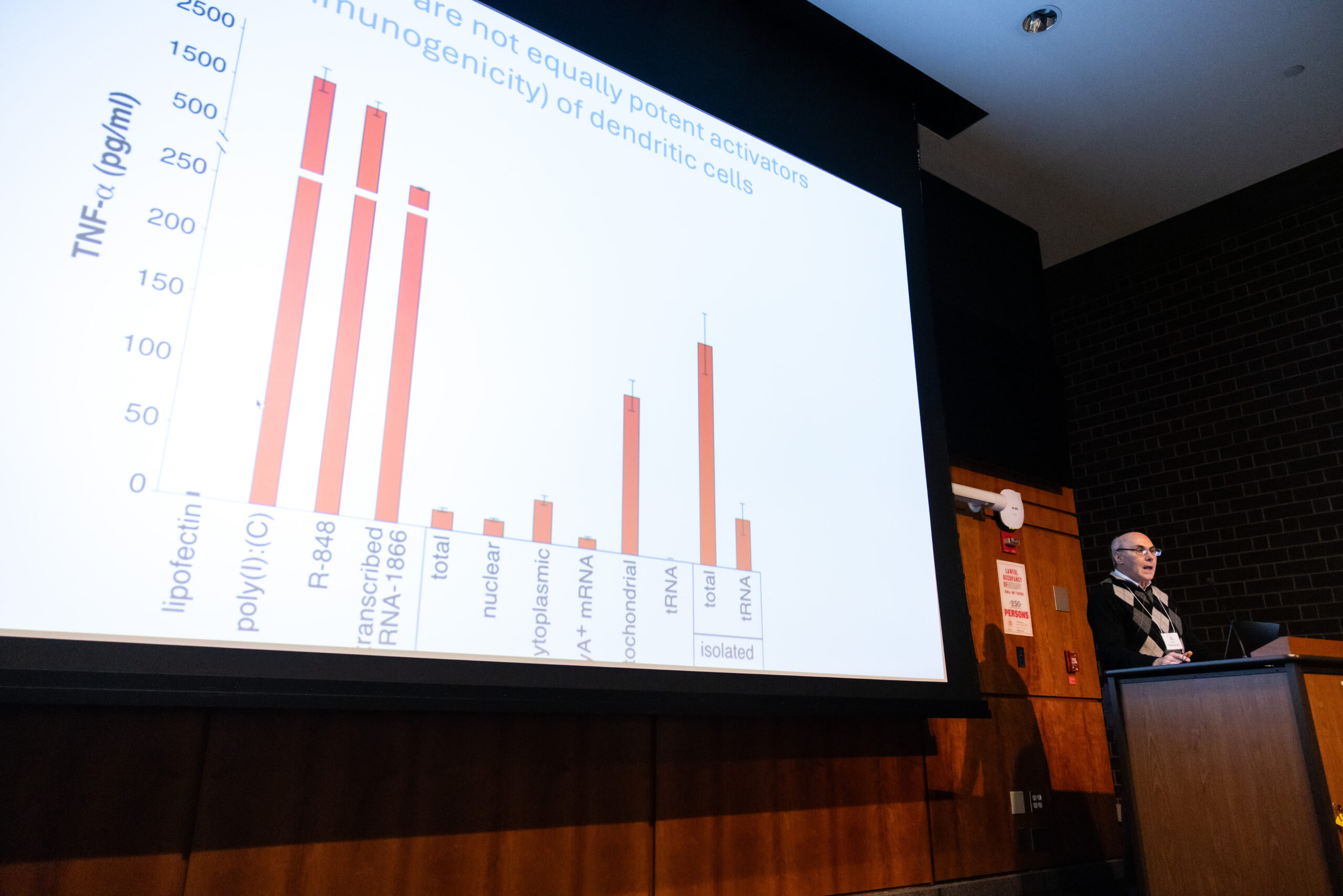
RNADay@Penn 2024
At the inaugural RNADay@Penn, RNA fans from around the country came to celebrate everything RNA! The day featured distinguished speakers, 30 RNA-based posters, entertainment, and food!
To be sure you are in the know for RNADay@Penn 2025, please join our membership list.
Welcome to the Penn Institute for RNA Innovation
The Penn Institute for RNA Innovation is dedicated to the understanding and development of all things RNA and will help form collaborations that will unify and link all elements from RNA-based basic science through therapeutic activities.
The Institute is led by Dr. Drew Weissman, Roberts Family Professor in Vaccine Research and Director of the Penn Institute for RNA Innovation. He is a Professor in the Department of Medicine and Director of Vaccine Research in the Infectious Diseases Division at the Perelman School of Medicine at the University of Pennsylvania.
Penn Institute for RNA Innovation Spring 2024 Pilot Grant Program
The Penn Institute for RNA Innovation at the University of Pennsylvania, CHOP, and Wistar seek to support RNA-related basic, translational and clinical research. Towards that goal, the Institute is requesting applications for its Pilot Grants Program for Spring 2024. Pilot funding will provide initial support to establish proof of concept or extend findings to enable extramural funding and publications in the future. Priority areas, deadlines, and application guidelines are provided here.
Penn Institute for RNA Innovation RNA Travel Grant Program
The Institute for RNA Innovation at the University of Pennsylvania, CHOP, and Wistar seeks to support RNA biology research to enhance basic science, translational, and clinical studies.
Travel Grants support graduate students and postdoctoral researchers who wish to attend and/or present their research at scholarly meetings off campus and may be used to defray the cost of travel and fees.
Click here to learn more.
2023 Nobel Prize Lectures with Drs. Kariko and Weissman
ABOUT
Dr. Drew Weissman M.D., Ph.D.
Dr. Drew Weissman is the Roberts Family Professor in Vaccine Research and Director of the Penn Institute for RNA Innovation. He is a Professor in the Department of Medicine and Director of Vaccine Research in the Infectious Diseases Division at the Perelman School of Medicine at the University of Pennsylvania.
Dr. Weissman, M.D., Ph.D., is a physician- scientist and pioneer in the science of immunology, with major contributions to the field. Notably, Dr. Weissman, and his colleagues discovered a novel nucleoside-modified mRNA platform that bypasses adverse immunologic response. The result of decades-long dedication and research, this platform now serves as the basis for the burgeoning research of targeted therapeutics for some of the world’s most devastating diseases. This research outcome paved the way for the first mRNA vaccines, being the critical backbone to the COVID-19 mRNA vaccines. Dr. Weissman and his lab team continue to conduct basic science research to understand and develop new nucleoside-modified mRNA platforms to advance effective and safe vaccines for different types of diseases, as well as new therapeutics.
Dr. Weissman is the recipient of multiple accolades in medicine and research, including the Lasker-DeBakey Award, the Breakthrough Prize, The Horwitz Prize and the Warren Alpert Foundation Prize.