Research Program
The skin is a formidable barrier and the first line of defense against the external environment. The skin is also colonized with microbial communities, which we hypothesize have critical functions in cutaneous health and disease. These functions may include but are not limited to colonization resistance to block invasion by opportunistic or pathogenic microbiota and regulation of immunity and inflammation. Various cutaneous disorders and infections are hypothesized to in part result from dysregulation of microbiota-host interactions. The Grice lab applies a variety of different approaches, model systems, and human tissues and isolates to elucidate mechanisms regulating such interactions.
Currently, research in the laboratory is divided into the following focus areas:
- (1) Skin microbiome regulation of epithelial defense
- (2) Microbial regulation of tissue repair and inflammation in chronic wounds
- (3) Skin microbiota-driven mechanisms of colonization resistance to pathogens
(1) Skin microbiome regulation of epithelial defense
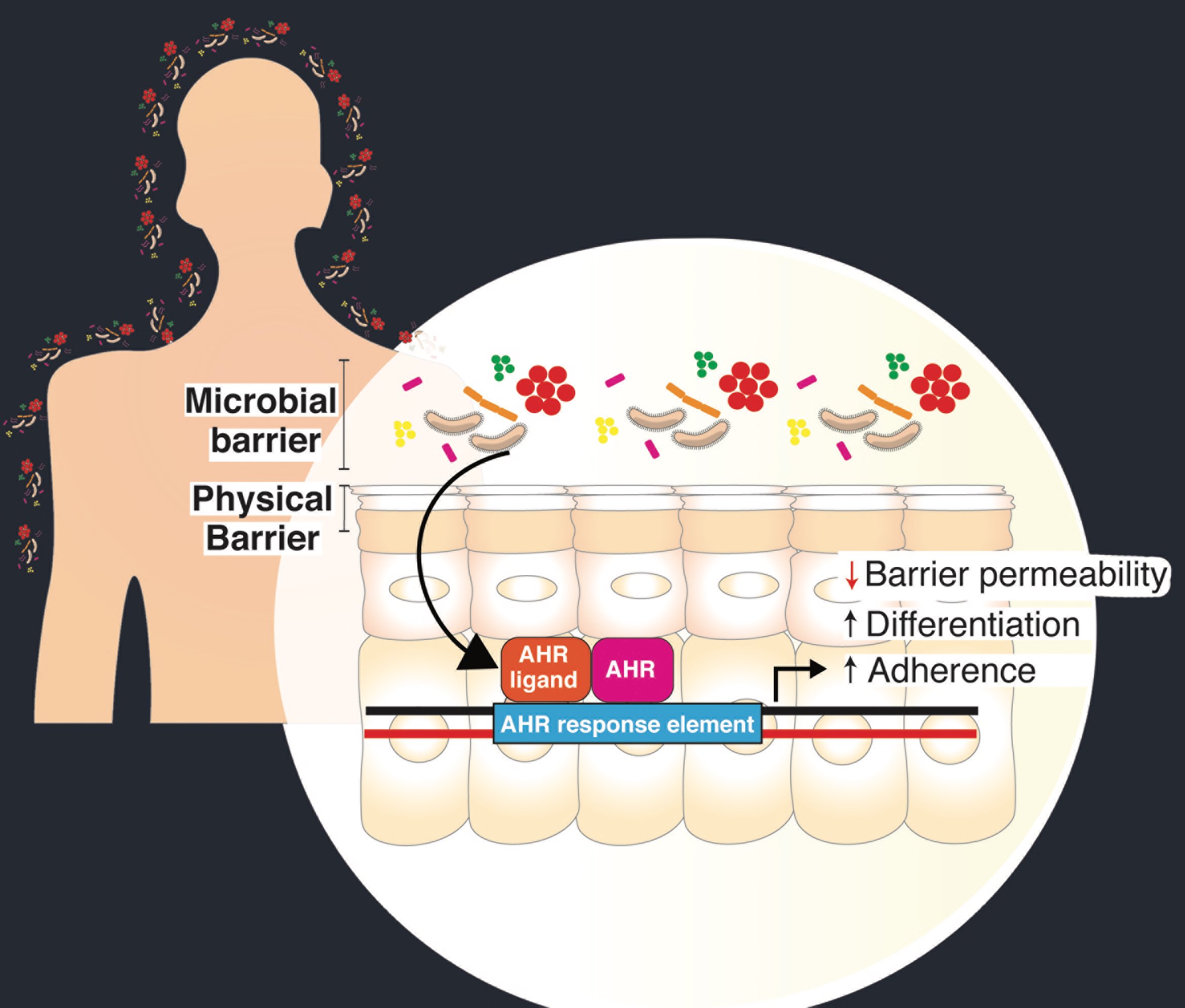
The skin microbiome has been associated with numerous inflammatory and barrier disorders, but the roles and mechanisms in regulating homeostatic barrier function is a major gap in our understanding of cutaneous host-microbe interactions. Using germ-free mice, we found that there are subsets of genes that are microbially regulated in the epidermis that are involved with epithelial development and differentiation, innate immunity, among others. These findings provided the basis to further examine molecular and biochemical mechanisms whereby the skin microbiota interact with the host. The lab’s most recent work implicates the keratinocyte aryl hydrocarbon receptor (AHR) by which human skin microbes regulate homeostatic epidermal permeability barrier formation, function, and defense against infection (Uberoi et al., 2021).
(2) Microbial regulation of tissue repair and inflammation in chronic wounds
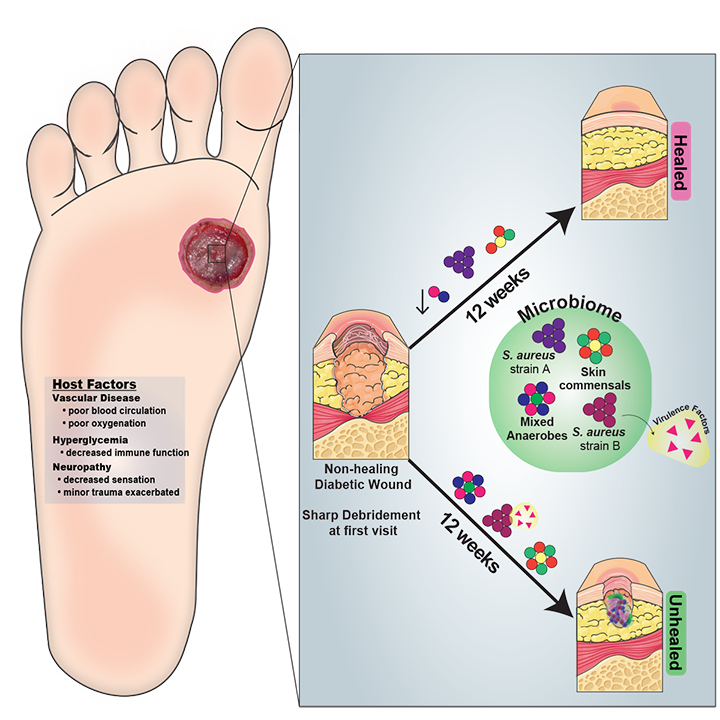
Chronic, non-healing skin wounds affect >6 million patients in the US and exceed $10 billion in health care costs annually. The wound’s microbial milieu is believed to play a key role in mediating healing outcomes, the development of complication, and/or response to therapy. We integrate the study of clinical wound specimens with basic research to elucidate the role of microbial communities in chronic wound outcomes. Our wound studies are currently focused on two different wound types and cohorts:
In collaboration with Dr. Sue Gardner at University of Iowa, we study diabetic foot ulcers and the wound microbiomes. Using 16S ribosomal RNA sequencing, we established patterns of microbial colonization associated with ulcer and patient characteristics as well as patient outcomes. This included the observation that microbial communities are more stable in wounds with poor outcomes (Loesche et al, 2017). We also employed shotgun metagenomic sequencing coupled with microbial genome sequencing to reveal that specific strains of S. aureus were associated with poor outcomes (Kalan et al, 2019). Current work utilizes a culture collection of wound isolates, genome sequencing, and ex vivo and in vivo wound models to identify and characterize microbial pathogenicity mechanisms associated with poor patient outcomes.
In collaboration with Dr. Phillip Scott at Penn Vet and Dr. Edgar Carvalho in Brazil, we study the role of skin microbiome in cutaneous leishmaniasis. We previously found that these non-healing lesions in humans are associated with alterations in the skin microbiome. Taking these findings to mouse models of leishmania infection, we demonstrated that dysbiotic skin microbiota enhanced lesion pathology. We are currently investigating how different types of microbial dysbiosis as well as strain-level variation in S. aureus species influence response to treatment, wound healing, and pathoinflammation.
(3) Skin microbiota-driven mechanisms of colonization resistance to pathogens
An exciting recent interest of the Grice lab is the study of competitive interactions in the skin microbiome, with the goal of leveraging these interactions in the prevention of colonization and/or treatment of infection. This work arose from early studies to test how perturbations to the skin microbiome influence colonization resistance to pathogens such as methicillin-resistant S. aureus (MRSA). In murine models and human subjects, topical antimicrobial treatments remove commensal skin microbes, which causes increased colonization efficiency of S. aureus on the skin. Building on these findings, ongoing investigations employ porcine skin models and microbial communities as a novel source of chemical diversity for the inhibition of MRSA. Combining high-throughput screens with biochemical and genomic approaches, we are identifying the determinants that confer colonization resistance to MRSA. Additionally, we are studying community assembly, succession, and function of transplanted synthetic communities in the host to understand microbial and host mechanisms that contribute to the engraftment of live microbial communities on the skin.