Our Interests
Bunyaviruses
Bunyavirales is an order of single-strand, spherical, enveloped RNA viruses with over 400 members. A significant number of these viruses are highly pathogenic in humans, and cause a wide range of severe diseases. Our projects focus on seven viruses from four families and genera in the Bunyavirales order.
We are in the process of developing recombinant VSV vaccines for two bandaviruses. Severe Fever with Thrombocytopenia Syndrome Virus (SFTSV), recently renamed Dabie bandavirus, is a tick-borne virus endemic to Eastern Asia that causes a hemorrhagic febrile illness. Heartland virus (HRTV), another tick-borne virus, emerged in Missouri, and has since caused cases of acute febrile illness throughout the Midwest and Southern United States. SFTSV and HRTV pose a threat to other parts of the world as the ticks that transmit them invade other regions. For example, the Asian Longhorned tick, which transmits SFTSV, is now an invasive species in the Eastern United States. Therefore, SFTSV has the potential to become endemic to the U.S. Both SFTSV and HRTV are classified as Category C priority pathogens by the National Institute of Allergy and Infectious Diseases (NIAID).
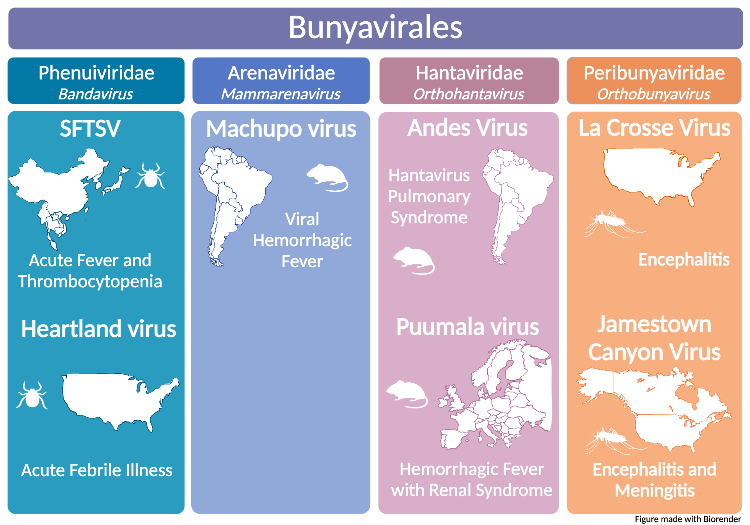
Machupo virus (MACV), the causative agent of Bolivian Hemorrhagic Fever, is a reemerging New World Arenavirus endemic to Bolivia. It is spread through the inhalation of mouse urine and feces. MACV is classified as a Category A priority pathogen because it poses such a significant risk to public health.
Andes virus (ANDV) and Puumala virus (PUUV) are two hantaviruses of interest to our lab. ANDV is a New World hantavirus found in South America that causes Hantavirus Pulmonary Syndrome (HPS), a severe life-threatening respiratory illness. Like MACV, it is listed by NIAID as a Category A priority pathogen. PUUV, an Old World hantavirus endemic to Scandinavia, Western Europe, and Russia, causes Hemorrhagic Fever with Renal Syndrome (HFRS). Like SFTSV and HRTV, PUUV is a Category C priority pathogen.
Our work on orthobunyaviruses centers on the California Serogroup (CSG) viruses, with particular emphasis on La Cross Virus (LACV) and Jamestown Canyon Virus (JCV). Both viruses are spread by mosquitos, are endemic to North America, and can cause encephalitis and meningitis. We aim to understand the genomic factors that influence neurovirulence and neuroinvasiveness in CSG viruses.
Recombinant VSV Vaccines
VSV is a highly cytopathic virus that has been developed as a vaccine vector due to its ability to induce strong, protective antibody and T-cell responses to encoded foreign antigens after a single dose. Although VSV can infect humans, it is not known to be a significant human pathogen, and unlike some other viral vaccine vectors, there is no significant preexisting immunity to VSV in humans. Licensing and implementing a recombinant VSV expressing the Ebola glycoproteins as a highly efficacious vaccine against Ebola highlights the potential of this platform.
To create recombinant VSV (rVSV), the native VSV glycoprotein gene (G) is replaced by a gene encoding a foreign viral glycoprotein. This replacement generates a replicating virus that mimics the foreign virus through expression of its glycoprotein. These recombinant viruses are replication competent, and can be used as viral vaccines or as a way to study BSL3 /BSL4 pathogens in a BSL2. The Bates lab has produced a number of recombinant VSV expressing bunyaviral and filoviral glycoproteins for work on vaccines and viral trafficking studies.
In parallel, we have generated nucleoside-modified mRNA encoding bunyaviral surface glycoproteins and produced lipid nanoparticle:mRNA vaccines. We are employing rVSV and LNP:mRNA alone and in combinations as candidate vaccines, and analyzing the induced humoral and cellular responses needed for protection in animal models. Currently, we are working on vaccines for SFTSV, Heartland, and Machupo viruses.
Virus Host Interactions
We use unbiased whole genome interrogation methods to define factors and processes important for viral infection. This research is based on the hypothesis that there are likely specific requirements that map to individual viruses or genera and common virus family factors and pathways that can be elucidated by comparative studies.
We have used whole genome and targeted screening methods to identify and characterize host factors needed for entry into host cells by the glycoproteins of several viral families including bunyaviruses (Hantaan, Andes, Sin Nombre, Severe Fever with Thrombocytopenia Virus and Heartland), filoviruses (Ebolavirus and Marburgvirus) and coronaviruses (SARS-CoV and SARS-CoV-2). Additionally, we are analyzing how viral glycoproteins are trafficked and processed by host cells.