The Integration of Mitochondrial and Mechanical Function
Traditional biologists tends to focus on metabolic origins and manifestations of disease, while pathological alterations in cell mechanics are of increasing importance to bioengineers. Many pathologies show concurrent dysfunction in both areas. For example, cancer cells famously prefer glycolysis in the well-known Warburg effect, and are also softer and weaker than normal cells. This work strives to connect these two areas, highlighting the intersection of mitochondrial and mechanical dysfunction in cells. Together with collaborators, we have pursued this topic on several fronts, investigating how mitochondrial dysfunction affects cell mechanics and the cytoskeleton, and vice versa. This research has also led to the development of several new tools that we are using in the course of our work.
1. How do changes in mitochondrial function affect cell mechanics and the cytoskeleton?
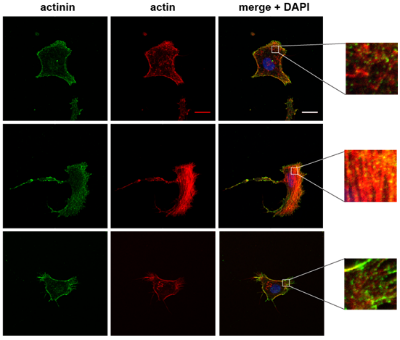
Mitochondrial dysfunction can occur due to a genetic defect or pharmacological inhibition, both of which we are studying in relation to cellular mechanics. In collaboration with members from Doug Wallace's lab at CHOP, we recently demonstrated that changes in heteroplasmy of a particular mitochondrial genetic defect manifest as cytoskeletal changes as well as alterations in cell mechanics. The resulting manuscript is in review, but some sneak peaks are provided below. The first image shows actin and actinin staining of cells harboring different heteroplasmy percentages, while the right image shows an atomic force microscopy (AFM) tip engaging with a cell to measure its elastic modulus via force spectroscopy.

2. How do cytoskeletal changes affect mitochondrial function?
Mitochondrial motility
As part of analyzing how cytoskeletal dysfunction affects mitochondria, we developed a tracking tool using ImageJ and Matlab to analyze mitochondrial motility on the whole cell level. Our methods are freely available online in Github, and along with our results, were recently published in Biotechnology and Bioengineering. This work was published as a spotlight by the journal, and showcased on the website as video highlight, as seen below:
Additionally, our paper was profiled by Penn Medicine which was highlighted by Penn News Today.
We first transfected human dermal fibroblasts to express fluorescent mitochondria, and then used ImageJ to pre-process images in order to view white mitochondria migrating against a black background. An original (left) and pre-processed (right) video (sped up 60x, so that one minute in real time is one second in the videos) are shown below.
We then used our algorithm to show that mitochondrial net distances in a given cell are lognormally distributed (below, left), as opposed to belonging to distinct categories of "diffusive" or "motor-driven." We further demonstrated that nocodazole, which depolymerizes microtubules, shifts this entire distribution towards lower net distances, while cytochalasin D, which depolymerizes actin filaments, shifted the distribution towards longer net distances (below, right). This indicates an important role for actin in mitochondrial motility, which may be cell-type dependent.
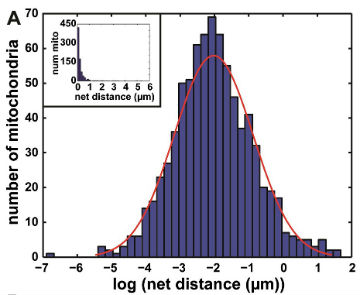
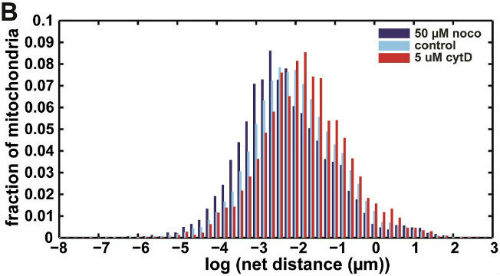
Further work we are conducting considers the effect of disrupted cell mechanics on other mitochondrial parameters.
3. Development of new tools and methods for analysis of cell mechanics and the cytoskeleton
Analysis of actin stress fiber orientation
We are working on a method to quantify orientation of actin stress fibers. This will be helpful our work, to demonstrate cytoskeletal effects of mitochondrial perturbation, but should also be highly valuable to bioengineers and others interested in actin stress fiber orientation. Below we highlight how we process images (left) and a rose-plot for that same cell illustrating actin fiber orientation (right).
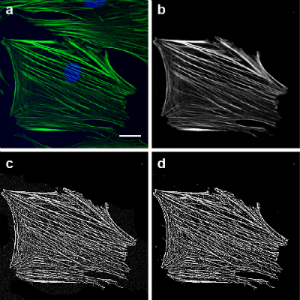
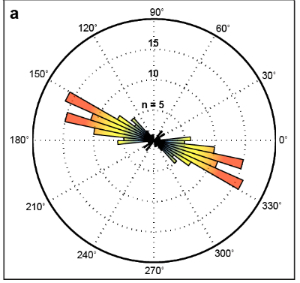
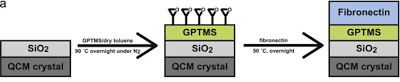
Development of chemically coated fibronectin surfaces for QCM-D cell experimentation
We are interested in using the quartz crystal microbalance with dissipation (QCM-D) for analyzing changes in cell mechanics. This method can provide mechanical information which is complementary to more traditionally used atomic force microscopy. To that end, we developed a methodology for chemically coating QCM-D sensors with fibronectin (chemFN) to optimize detection of changes in cell mechanics. This work was published in Biosensors and Bioelectronics in 2014.
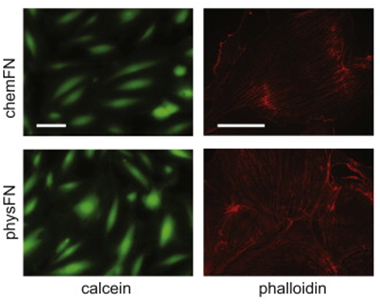
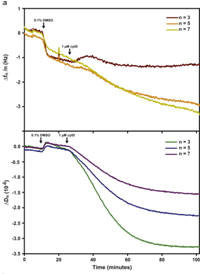
An overview of the chemical synthesis is shown below on the top left, while the figure on the bottom left shows that cells exhibit comparable calcein and phalloidin staining on chemFN surfaces to those on traditionally coated physisorbed fibronectin surfaces. The right image shows the change in dissipation we observed when cells plated on a chemFN-coated QCM-D sensor were subjected to cytochalasin D.