Greenberg Lab Research
News
We're excited to share the following news updates:
- A big congratulations to Dr. Sangin Kim for being awarded a Damon Runyon Postdoctoral Fellowship!
- Commendations to Dr. Tao Shi for receiving the competitive Basser Postdoctoral Fellowship Award!
- Congratulations to co-authors Drs. Haoyang Jiang and Tianpeng Zhang, for their publication in Molecular Cell that was featured on the cover:
- Jiang H*, Zhang T*, Kaur H, Shi T, Krishnan A, Kwon Y, Sung P, Greenberg RA. BLM helicase unwinds lagging strand substrates to assemble the ALT telomere damage response. Molecular Cell 2024 May 2;84(9):1684-1698.e9. doi: 10.1016/j.molcel.2024.03.011. Epub 2024 Apr 8. *Co-First Authors
- Collaboration between PCGI Director Roger Greenberg and Elton Zeqiraj (Univ. of Leeds) reports a new mechanism that ensures replication fidelity. Congratulations to first authors Vidhya Krishnamoorthy (Greenberg Lab, UPENN), Martina Foglizzo (Leeds), Robert Dilley (Greenberg Lab alum). Featured in Penn Today news April 4, 2024: Krishnamoorthy V, Foglizzo M, Dilley RL, Wu A, Datta A, Dutta P, Campbell LJ, Degtjarik O, Musgrove LJ, Calabrese AN, Zeqiraj E, Greenberg RA. The SPATA5-SPATA5L1 ATPase complex directs replisome proteostasis to ensure genome integrity. Cell 2024 Mar 22;S0092-8674(24)00250-2.doi: 10.1016/j.cell.2024.03.002. Online ahead of print.
- Congratulations to Tianpeng Zhang on his new position as Assistant Professor at the University of Virginia Medical School
- Congratulations to Haoyang Jiang on his Postdoctoral Fellowship award from the American Cancer Society
Germline mutations to the Breast Cancer 1 (BRCA1) or Breast Cancer 2 (BRCA2) genes are the major cause of hereditary breast and ovarian cancer susceptibility. Clinical BRCA1 and BRCA2 mutations render cells deficient in error-free mechanisms of DNA repair known as homologous recombination, implicating these activities in tumor suppression and response to genotoxic therapies.
Our work has revealed a molecular understanding for how BRCA1 recognizes DNA damage and competes with opposing DNA repair proteins to control genome integrity. We have demonstrated that an interaction between the BRCA1 BRCT domain and the RAP80 ubiquitin binding protein targets BRCA1 to K63-linked ubiquitin structures present at DNA damage sites. RAP80 ubiquitin interaction motifs (UIMs) provide a ubiquitin recognition element to target BRCA1 and a K63-ubiquitin specific deubiquitinating enzyme BRCC36 to DNA double strand breaks. Each of these activities is required for appropriate DNA damage checkpoint and repair responses (Sobhian et al. Science 2007; Shao et al. Genes&Dev 2009; Tang et al. Nat Struct & Mol Biol 2013; Jiang et al. Genes Dev 2015; Zeqiraj et al. Mol Cell 2015). Cancer causing BRCA1 BRCT mutants fail to interact with RAP80 and consequently demonstrate inefficient recruitment to DNA damage sites. Moreover, germline mutations in RAP80 and Abraxas are present in familial breast cancer (Nikkila et al. Oncogene 2009; Solyom et al Sci Transl Med 2012) and biallelic BRCA1 mutations cause a new subtype of Fanconi Anemia known as Fanc-S (Domchek et al. Cancer Discov 2013; Sawyer et al Cancer Discov 2015). Thus, a series of ordered events involving ubiquitin recognition, breakdown and synthesis are required for BRCA1-dependent DNA damage responses and tumor suppression.
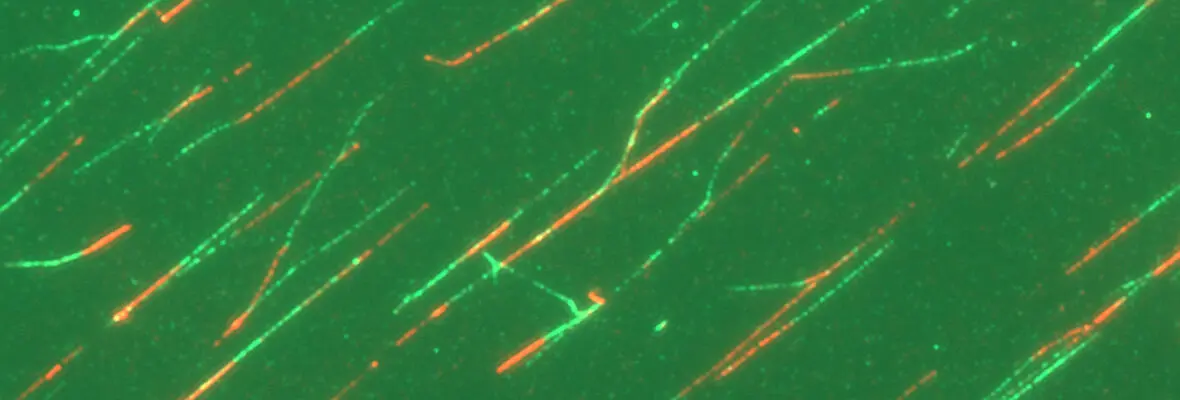
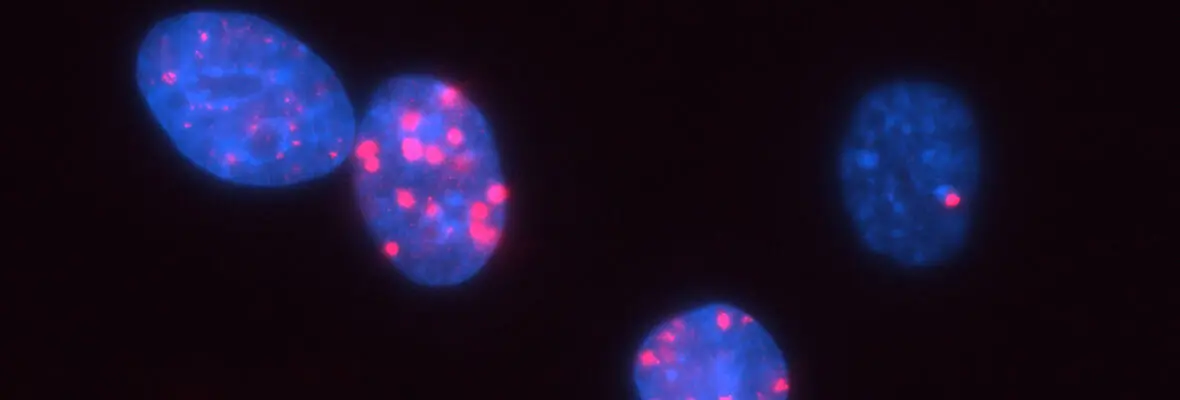
A second area of interest is the complex relationship between chromatin structure and DNA repair. We have developed several novel systems to investigate interrelationships between chromatin structure and DNA double strand break (DSB) repair. This was instrumental to our discoveries that DSBs silence transcription for multiple kilobases of chromatin in cis to the site of DNA damage (Shanbhag et al. Cell 2010), and that chromatin environment affects DNA repair mechanism choice and sensitivity to PARP inhibitors (Tang et al. Nat Struct Mol Biol 2013). More recently, we have developed methodologies to directly monitor homologous recombination at telomeres, a first for any genomic location in mammalian cells. This enabled our discovery of a novel form of homology directed repair that is responsible for alternative telomere length maintenance mechanisms in approximately 15% of human cancers (Cho et al. Cell 2014; Dilley et al. Nature 2016; Verma et al. Genes Dev 2019).
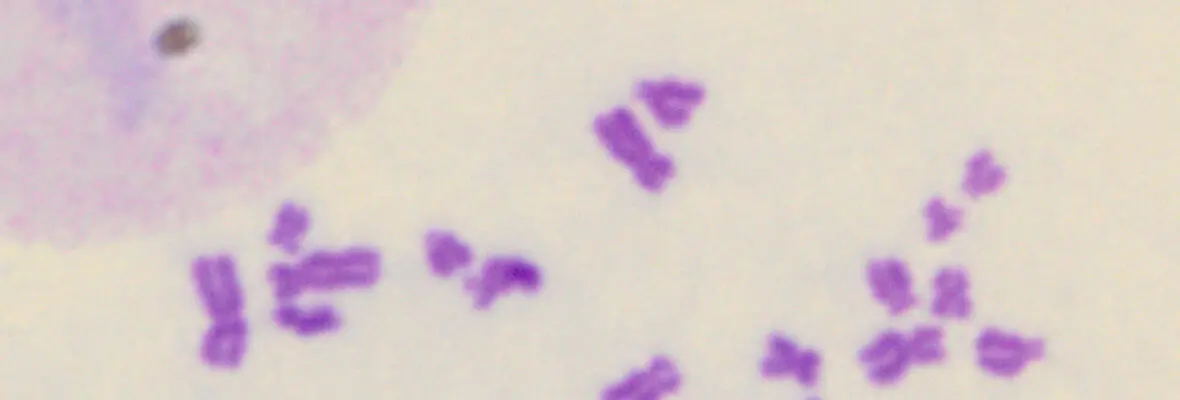
In addition to these studies involving acute DNA damage responses, we have recently determined the basis for the longstanding observation that DNA damage activates innate and adaptive immune responses (Harding et al. Nature 2017; Walden, Tian et al. Nature 2019). Our findings reveal that mitotic progression in the presence of DNA damage results in micronuclei within the cytoplasm that are recognized by the pattern recognition receptor cGAS. This produces inflammatory cytokine signals that activate anti-tumor immune responses to eradicate cells within the primary tumor and distal metastases. We will continue to use these systems to determine how DNA damage response mechanisms contributes to genome integrity, cancer etiology and response to therapy.
Perelman School of Medicine Events
-
PCGI Trainee Seminar
Monday, April 25, 2022
4:00-5:00 PM
Timothy Lippert (Greenberg Lab) presenting “DNA damage-driven inflammasome activation in BRCA-deficient ovarian cancer”
and
Jon Acosta (Feldser Lab) presenting “Elucidating the molecular determinants of p53-mediated pleiotropic effects in SCLC”