Cancer Imaging Phenomics
Cancer Imaging Phenomics: Radiomics, Radiogenomics, and Beyond
The lab has a great deal of activity in the field of computational oncology, spanning across various anatomies with primary emphasis on brain gliomas and lung cancer. Current focus is to provide precision diagnostics by characterizing tumor heterogeneity via machine learning based imaging signatures, thereby leading to optimized, personalized cancer treatment plans, as well as to develop tools for computational pathology and integrated diagnostics.
Current Projects
-- Precision diagnostics in glioblastoma.
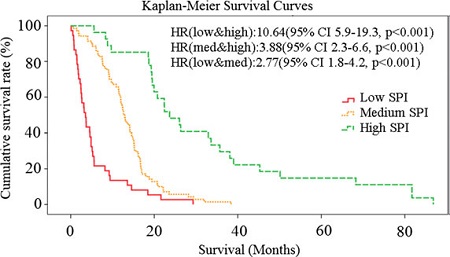
The phenotypic characteristics of GBM vary tremendously across patients, thereby indicating that this is not a single disease. Machine learning has shown promise in characterizing this heterogeneity, thereby allowing us to detect imaging signatures that relate to tumor subtype and clinical outcome. Examples of our work include the discovery of 3 subtypes of GBM, which had different prognosis as well as different molecular composition [1]; the development of a predictor of patient survival after standard of care [2]; the development of an imaging signature differentiating between pseudoprogression due to treatment, and true progressive disease [3, 4]. An example of survival curves according to baseline predictions are shown below
Predictive maps of peri-tumoral infiltration beyond resection margins.
We have developed predictive models of peri-tumoral infiltration in edematous regions, thereby highlighting peri-tumoral heterogeneity and its relationship to tumor progression[5, 6]. The figure below shows two representative examples of a heat map, with red regions being more likely to present future recurrence, and the relative agreement with post-resection follow up scans (months later), with the enhancing regions representing the actual recurrence.
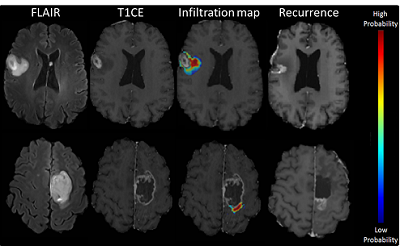
The overall goal of this line of research is to guide extensive, yet targeted peri-tumoral resection and focused radiation therapy in regions relatively more likely to recur early.
— Imaging Genomics (Radiogenomics)
The lab contributes to the rapidly expanding field of radiogenomics. Machine learning (both conventional and deep learning methods) have shown great promise in identifying complex imaging signatures that flag the presence of certain mutations. Importantly, this is done from standard clinical scans, rather than from specialized protocols involving radiotracers. Our laboratory has identified imaging signatures of EGFRvIII[7, 8] and IDH1[9] mutations in GBM, as well as of MGMT methylation[10]. Current work involves 49 mutations detected via a specialized next generation sequencing panel we have built specifically for GBM. Moreover, our group is extending these methods to better characterize regional heterogeneity of radiogenomic signatures, thereby aiming to estimate the “degree of expression” of different radiogenomic signatures throughout the tumor.
Publications
[1] S. Rathore et al., "Radiomic MRI signature reveals three distinct subtypes of glioblastoma with different clinical and molecular characteristics, offering prognostic value beyond IDH1," Nature Scientific Reports, vol. 8, no. 1, p. 5087, 2018/03/23 2018.
[2] L. Macyszyn et al., "Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques," Neuro-oncology, vol. 18, no. 3, pp. 417-25, Mar 2016.
[3] H. Akbari et al., "Quantitative radiomics and machine learning to distinguish true progression from pseudoprogression in patients with GBM," American Society of NeuroRadiology (ASNR) 56th Annual Meeting, 2018.
[4] S. R. H. Akbari, S. Bakas, M. Nasrallah, M .Rozycki, S. Mohan, R. Wolf, M. Bilello, M. Martinez-Lage, C. Davatzikos, "Quantitative image analysis and machine learning techniques for distinguishing true progression from pseudoprogression in patients with glioblastoma," presented at the the 23rd Annual Scientific Meeting of the Society for Neuro-Oncology (SNO), New Orleans, LA, November 15 - 18, 2018, 2018.
[5] H. Akbari et al., "Imaging Surrogates of Infiltration Obtained Via Multiparametric Imaging Pattern Analysis Predict Subsequent Location of Recurrence of Glioblastoma," Neurosurgery, vol. 78, no. 4, pp. 572-80, Apr 2016.
[6] S. Rathore et al., "Radiomic signature of infiltration in peritumoral edema predicts subsequent recurrence in glioblastoma: implications for personalized radiotherapy planning," J Med Imaging (Bellingham), vol. 5, no. 2, p. 021219, Apr 2018.
[7] H. Akbari et al., "In vivo evaluation of EGFRvIII mutation in primary glioblastoma patients via complex multiparametric MRI signature," Neuro-oncology, vol. 20, no. 8, pp. 1068-1079, Jul 5 2018.
[8] S. Bakas et al., "In Vivo Detection of EGFRvIII in Glioblastoma via Perfusion Magnetic Resonance Imaging Signature Consistent with Deep Peritumoral Infiltration: The phi-Index," Clinical cancer research : an official journal of the American Association for Cancer Research, vol. 23, no. 16, pp. 4724-4734, Aug 15 2017.
[9] Spyridon Bakas et al., "Non-Invasive In Vivo Signature of IDH1 Mutational Status in High Grade Glioma, From Clinically-Acquired Multi-Parametric Magnetic Resonance Imaging, Using Multivariate Machine Learning," Neuro-oncology, vol. 20, pp. vi184-vi185, 2018.
[10] S. Rathore et al., "Non-invasive determination of the O6-methylguanine-DNA-methyltransferase (MGMT) promoter methylation status in glioblastoma (GBM) using magnetic resonance imaging (MRI)," Journal of Clinical Oncology, vol. 36, no. 15_suppl, pp. 2051-2051, 2018.
coming soon

