Federated Learning in Medicine: Facilitating Multi-Institutional Collaboration Without Sharing Patient Data
Federated learning (FL) was introduced by Google in 2017 and describes a distributed machine learning framework enabling multi-institutional collaborations without sharing data among the collaborators.
In September 2018, in collaboration with Intel AI, we presented the results of the first application of FL to real-world medical imaging data, during the International Medical Image Computing and Computer Assisted Interventions (MICCAI) conference in Granada, Spain (Fig.1). This initial study of ours, was published as a Springer’s Lecture Notes in Computer Science proceedings paper [1] denoting the first publication on FL in the medical domain. We specifically demonstrated that a deep learning model [6] trained using FL could reach 99% of the performance of the same model trained with the traditional data-sharing method [2-5].
In 2019, the Informatics Technology for Cancer Research (ITCR) program of the National Cancer Institute (NCI) of the National Institutes of Health (NIH), awarded a three-year $1.2M federal grant to CBICA (Principal Investigator: Spyridon Bakas), to develop a FL framework focusing on tumor segmentation, in collaboration with Intel AI. As part of this grant, we are currently leading the first real-world medical use case of FL, coordinating a federation of 29 collaborating institutions around the world.
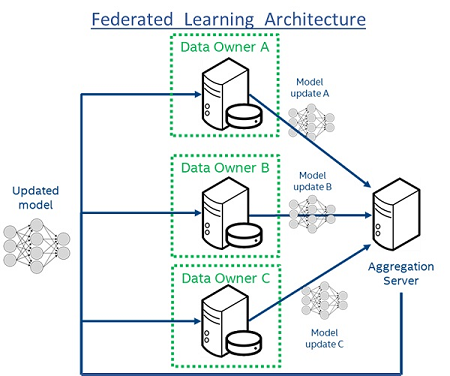
Figure 1: Federated Learning Architecture. Each “Data Owner” trains a model locally using only their own data. Each model is then shared with the central “Aggregation Server”, which creates a consensus model (“Updated model”) that has accumulated knowledge from all “Data Owners”. The raw data are always retained within the “Data Owners” never leaves the institutions, which not only adds privacy but also prevents large data transfers on the network.
- Sheller MJ, Reina GA, Edwards B, Martin J, Bakas S. Multi-institutional Deep Learning Modeling Without Sharing Patient Data: A Feasibility Study on Brain Tumor Segmentation. Lecture Notes in Computer Science book series (Volume 11383). 2019.
- Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby JS, Freymann JB, Farahani K, Davatzikos C. “Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features”, Nature Scientific Data, 4:170117 (2017) DOI: 10.1038/sdata.2017.117
- Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby J, Freymann J, Farahani K, Davatzikos C. “Segmentation Labels and Radiomic Features for the Pre-operative Scans of the TCGA-GBM collection”, The Cancer Imaging Archive, 2017. DOI: 10.7937/K9/TCIA.2017.KLXWJJ1Q
- Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby J, Freymann J, Farahani K, Davatzikos C. “Segmentation Labels and Radiomic Features for the Pre-operative Scans of the TCGA-LGG collection”, The Cancer Imaging Archive, 2017. DOI: 10.7937/K9/TCIA.2017.GJQ7R0EF
- Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R, Lanczi L, Gerstner E, Weber MA, Arbel T, Avants BB, Ayache N, Buendia P, Collins DL, Cordier N, Corso JJ, Criminisi A, Das T, Delingette H, Demiralp Γ, Durst CR, Dojat M, Doyle S, Festa J, Forbes F, Geremia E, Glocker B, Golland P, Guo X, Hamamci A, Iftekharuddin KM, Jena R, John NM, Konukoglu E, Lashkari D, Mariz JA, Meier R, Pereira S, Precup D, Price SJ, Raviv TR, Reza SM, Ryan M, Sarikaya D, Schwartz L, Shin HC, Shotton J, Silva CA, Sousa N, Subbanna NK, Szekely G, Taylor TJ, Thomas OM, Tustison NJ, Unal G, Vasseur F, Wintermark M, Ye DH, Zhao L, Zhao B, Zikic D, Prastawa M, Reyes M, Van Leemput K. “The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS)”, IEEE Transactions on Medical Imaging 34(10), 1993-2024 (2015) DOI: 10.1109/TMI.2014.2377694
- Ronneberger O., Fischer P. , and T. Brox. “U-Net Convolutional Networks for Biomedical Image Segmentation.” arXiv:1505.04597v1 [cs.CV] 18 May 2015

