Individualized functional network modeling
Delineation of large-scale functional networks from resting state functional MRI data has become a standard tool to explore the functional brain organization in neuroscience. However, existing methods sacrifice subject specific variation in order to maintain the across-subject correspondence necessary for group-level analyses. In order to obtain subject specific functional networks that are comparable across subjects, we have developed data-driven methods for detecting subject specific functional networks while establishing group level correspondence, including a group information guided independent component analysis (GIG-ICA) [11] and a sparse non-negative matrix decomposition method [12]. The later has also been further extended to model subject-specific functional networks at multiple spatial scales with a hierarchical organization [13].
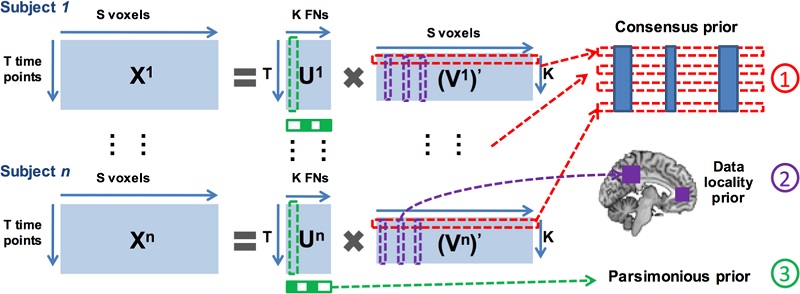
Schematic diagram of the sparse non-negative matrix decomposition method. The fMRI data of each individual subject are arranged as a matrix with each row for one time point and each column for one voxel. The fMRI data of a group of subjects are simultaneously decomposed into non-negative subject specific functional networks with their corresponding time courses in a collaborative setting with 3 regularization terms: 1) a voxel-wise group sparsity regularization term is adopted as an inter-subject consensus prior so that spatial correspondence and variations of functional networks of different subjects are preserved simultaneously; 2) a data locality regularization term is adopted to enhance both functional coherence and spatial proximity of voxels so that spatially continuous and functionally coherent voxels are encouraged to reside in the same functional network and 3) an intra-subject parsimonious regularization term is adopted to eliminate redundant functional networks with similar functional profiles using automatic relevance determination techniques.

