Research
Our research group has broad expertise in cryo-electron microscopy (cryo-EM), cross-linking mass spectrometry, and computational model building. We combine these cutting-edge structural techniques with biochemistry and genetics using S. cerevisiae as a model system, and focus on four main research areas, described below:
The goal of this research is to reconstitute the transition from transcription initiation to elongation from purified components from S. cerevisiae and to elucidate the mechanisms of positive and negative control. Transcription initiation can be reconstituted from highly purified six general transcription factors (GTFs), RNA polymerase II (pol II), and promoter DNA, that assemble the ~30-subunit pre-initiation complex (PIC). However, earlier biochemical reconstitution systems had a technical limitation, namely very poor initiation efficiency. Thus, how pol II in the PIC escapes promoter and transitions to productive elongation complex remains unknown. This process involves not only GTFs and pol II, but also a variety of transcription factors and chromatin factors including +1 nucleosome that generally occludes the transcription start site. Building on our initial success in developing highly efficient in vitro reconstitution from yeast at quality and quantity amenable to structure determination (Fujiwara and Murakami, Methods, 2019), our laboratory further improved our in vitro transcription system to at least ~70% efficiency per template (Fujiwara et al., PNAS, 2019). By then isolating and characterizing naturally generated post-initiation complexes, we found that the initially-transcribing complex (ITC) containing pol II, GTFs, and a nascent transcript, persists much longer than previously proposed, implicating at least two conserved elongation factors as active components of this transition (Fujiwara et al., PNAS, 2019), consistent with our earlier single-molecule optical trapping studies (Fazal et al., Nature, 2015). By imaging reaction intermediates in our in vitro system, we have resolved multiple cryo-EM structures of transition intermediates including GTFs/pol II and determined the transition pathway involving many steps for regulation (Yang et al., Mol. Cell, 2022). Recently, the formation of such a long-persisting ITC formation established in yeast was reproduced in the human system, becoming recognized as a widespread regulatory point in all eukaryotes.
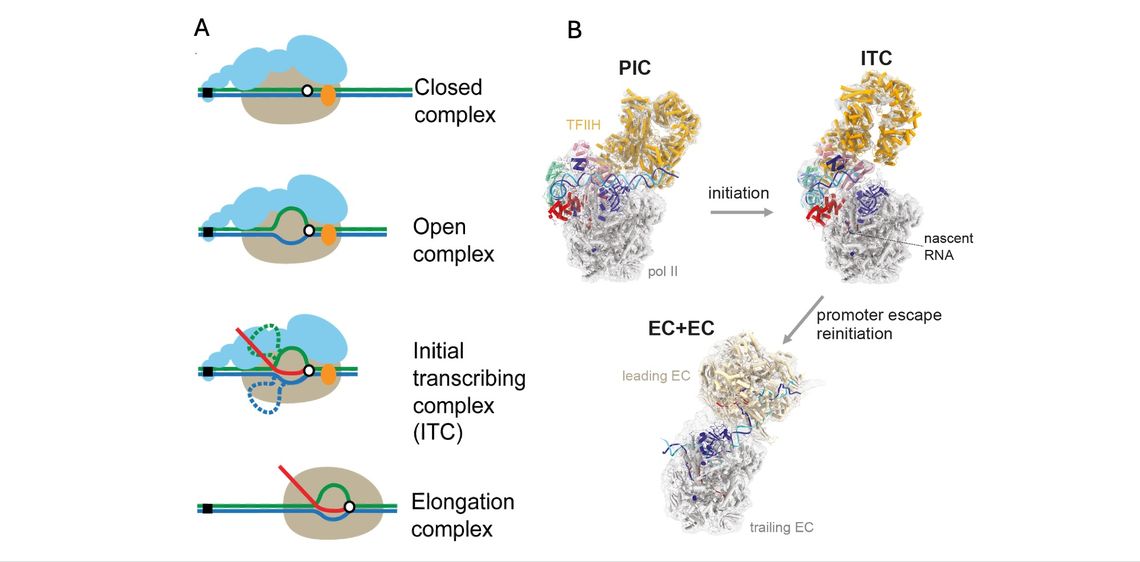
(B) The cryo-EM structure of the PIC, the ITC, and ECs colliding head-to-end (EC+EC) at 3.2-4.6 Å, 3.2-9.9 Å, and 3.5 Å resolutions (Yang et al., Mol. Cell, 2022).
We study the mechanism of chromatin assembly by the Hir/HIRA complex during active transcription with the Marmorstein lab at UPenn. The evolutionarily conserved HIRA/Hir histone chaperone complex and ASF1a/Asf1 co-chaperone cooperate to deposit histone (H3/H4)2 tetramers on DNA after transcribing RNA polymerase II displaces nucleosomes. The molecular architecture of the HIRA/Hir complex and its mode of histone deposition have remained unknown for more than three decades. Recently we have succeeded in determining the structure of the S. cerevisiae Hir complex with Asf1/H3/H4 through a combination of cryo-EM single-particle analysis and XL-MS (Kim et al., Mol. Cell, 2024). The structure suggests a first-of its kind model for how the Hir/Asf1 complex promotes formation of histone tetramers and their subsequent deposition onto DNA.
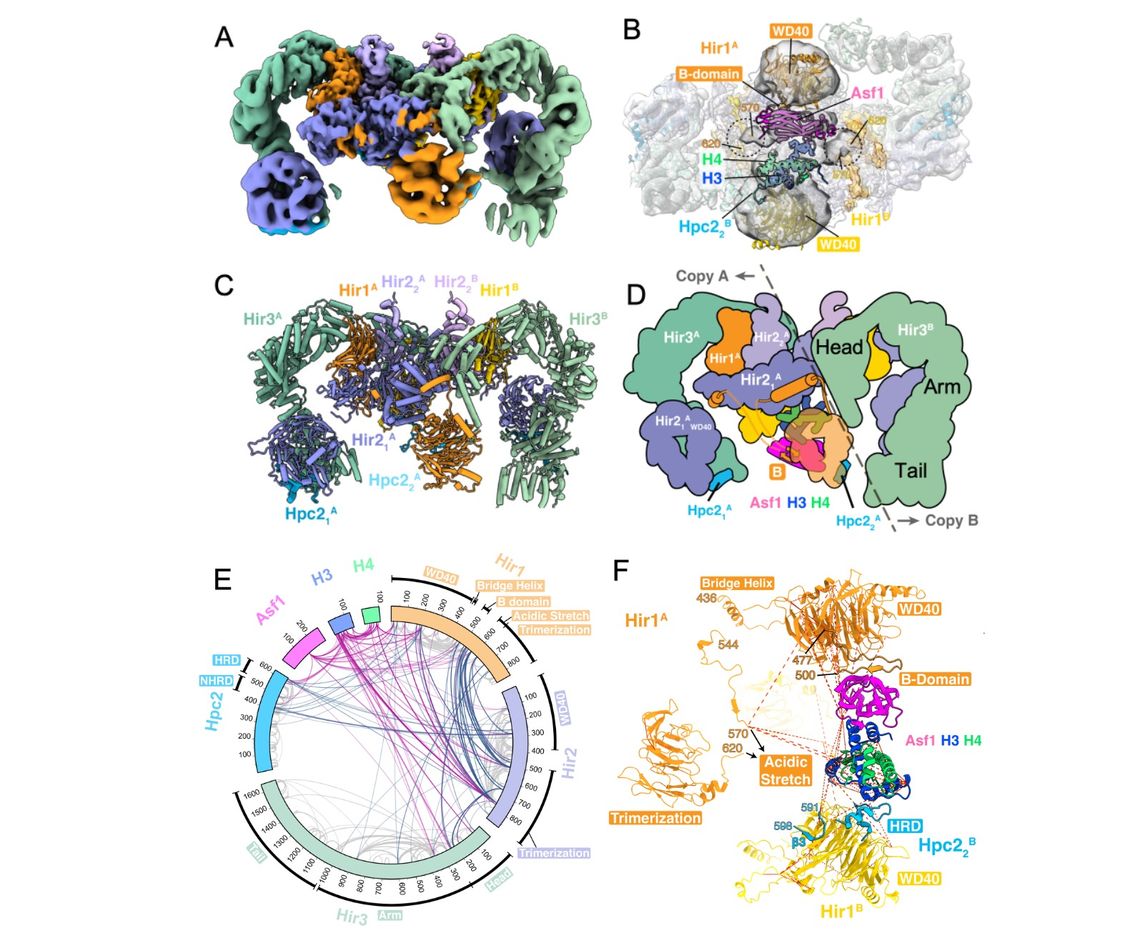
(A) Cryo-EM map of overall Hir structure bound to Asf1/H3/H4 at overall 2.96 Å resolution.
(B) One Asf1/H3/H4 complex was modeled into cryo-EM density at overall 8.3 Å resolution. Viewed from the bottom to see a substrate-binding “cavity”.
(C) Structure of the Hir complex corresponding to (A).
(D) Schematic representation of the complex.
(E) Crosslinking mass spectrometry (XL-MS) of the Hir complex. Circular plot of unique cross-linked residue pairs for the Hir complex (steel blue) and the Hir complex-Asf1/H3/H4 (violet red). Intra-subunit unique residue pairs are indicated in gray.
(F) Zoomed view of Asf1/H3/H4 in the cavity. A previous crystallographic model of Asf1/H3/H4 was docked into EM density using XL-MS as constraints. Unique cross-linked residue pairs identified by XL-MS are indicated by red dashed lines.
By extending our structural mechanistic studies of the PIC, we aim to reconstitute the complete process of transcription initiation in the context of chromatin from transcriptionally-repressed chromatin to the PIC assembly mediated by the Mediator co-activator complex. As a starting point for this undertaking, we employed cryo-EM single-particle analysis and cryo-electron tomography to resolve the Mediator-PIC on the GAL80 promoter with a cognate activator protein Gal4-VP16. The structure revealed two distinct Mediator-PICs in a dimeric form through the Mediator Tail module, induced by the homodimeric activator protein, consistent with the emerging picture of divergent transcription that arises from the majority of active pol II promoters in vivo. Furthermore, our structure reveals a critical component of functional significance, absent from previous structures – DNA extending from the point of contact with the Med15 subunit of the Mediator Tail, to about 80 bp of DNA upstream, reaching the second copy of the Mediator Tail. The structure of the PIC-Mediator in this novel form explains the requirement for Mediator for transcription activation and has opened new doors to understanding its role in divergent transcription (Gorbea et al., Mol. Cell, 2023).
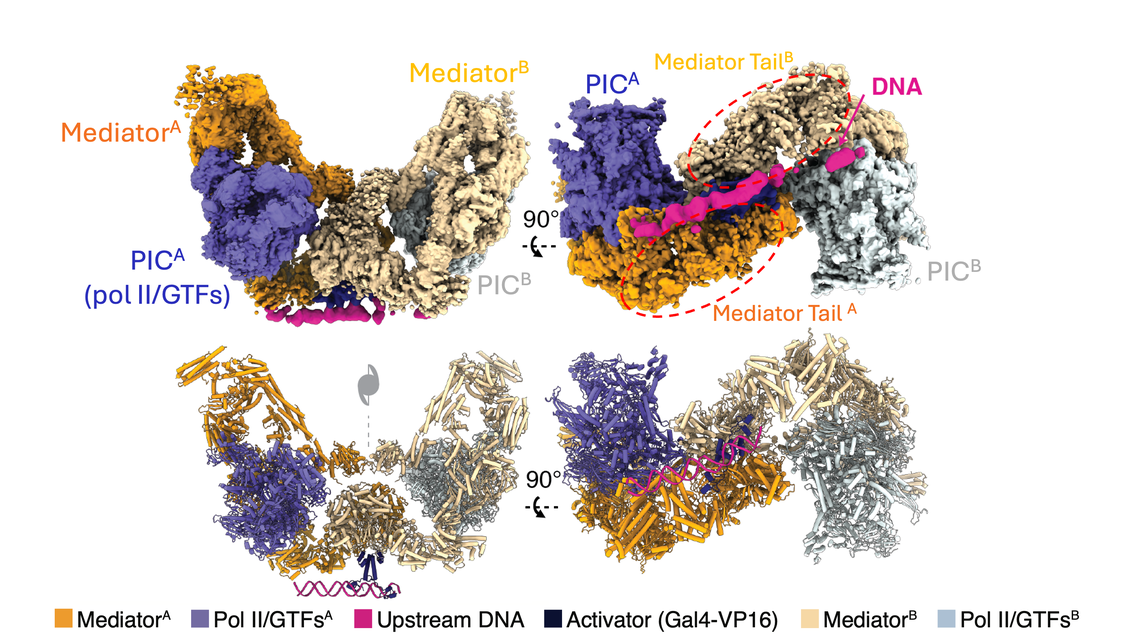
Lower panel: a model derived from cryo-EM density.
Promoter DNA is too flexible and averaged out during cryo-EM single-particle analysis (SPA). Capturing the comprehensive view including the path of the entire promoter DNA is critical in understanding promoter information transfer to the PIC. We are developing a hybrid approach of cryo-EM SPA and cryo-EM tomography that allows us to visualize the structural ensemble of the entire promoter DNA from a same specimen through a collaboration with the Chang lab at UPenn. While cryo-EM SPA reveals the structure core at near atomic resolution, cryo-ET allows us to see what subunits are bound to DNA within the complex, which region of the DNA are bound to the complex, and which region of the DNA loops out, all on a per-particle basis. This method demonstrated the value of the method that visualizes intermediates and heterogeneous populations that were not previously revealed by canonical cryo-EM SPA, and thus spur its application to a wide range of problems with other protein-DNA complexes (Gorbea et al., Mol. Cell, 2023).
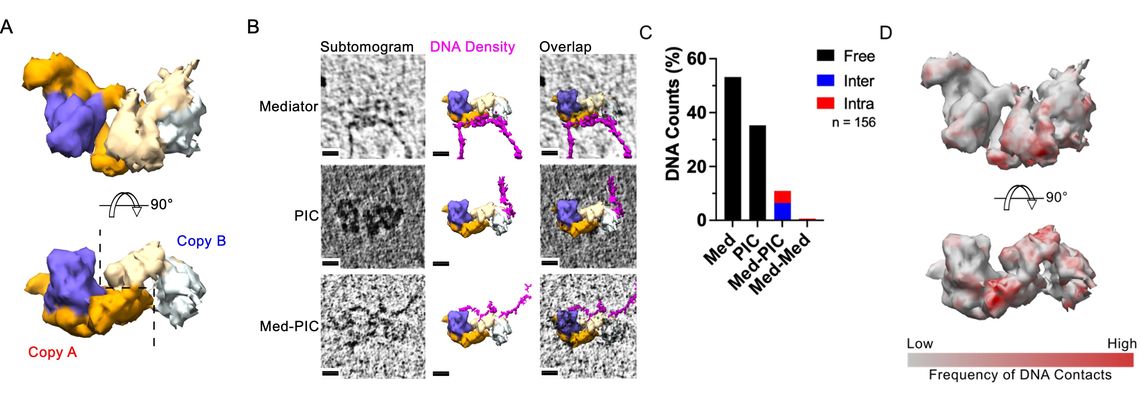
(A) Side view (Upper) and bottom view (Lower) of subtomogram averages.
(B) Raw individual subtomograms (left column), fitted subtomogram averages and segmented apparent DNA densities (middle column), and overlaps (right column). All viewed from the bottom. Scale bar: 10 nm.
(C) Relative population of different DNA trace classes based on the DNA contacts with the protein structures. Mediator: a DNA trace extending from the Mediator region; Med-PIC: a DNA trace connecting Mediator and PIC regions; PIC: a DNA trace extending from the PIC region; PIC-PIC: a DNA trace connecting the dimeric PIC regions.
(D) Side view (Upper) and bottom view (Lower) of heat map of DNA contacts on the subtomogram average.
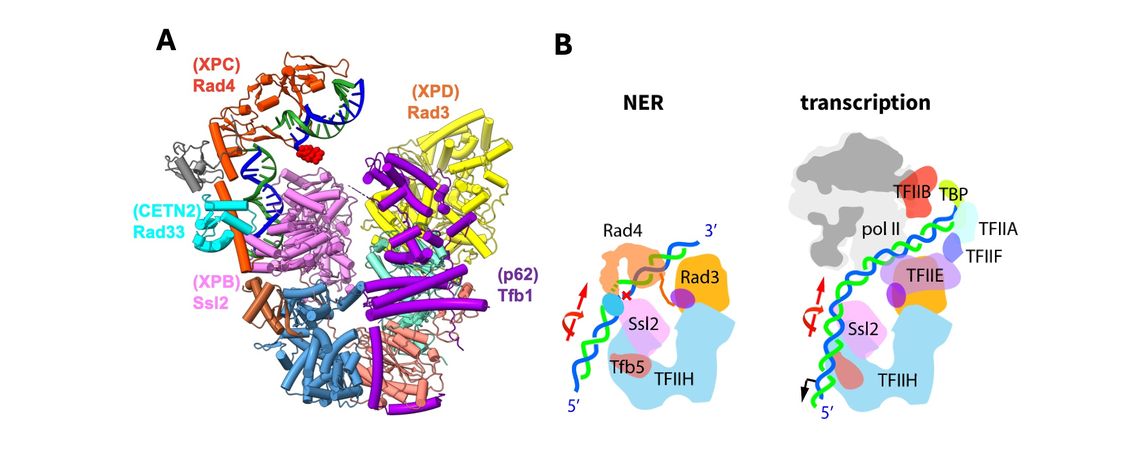
The goal of this research is to elucidate the dual function of TFIIH, the largest of the GTFs, in nucleotide excision repair (NER), a DNA repair pathway that acts as a guardian of genome integrity. The significance of failure of the NER system is highlighted by the rare cancer syndrome XP (xeroderma pigmentosum) in which the NER components are congenitally altered, and consequently, the incidence of skin cancers is increased ~10,000 fold. By employing cryo-EM and extensive XL-MS analyses, we have determined a structure of TFIIH complexed with Rad4-Rad23-Rad33 (the yeast homolog of XPC-RAD23B-CETN2) on a bona-fide damaged DNA through a collaboration with the Min lab (van Eeuwen et al., Nat. Commun, 2021). This “NER initiation complex” has for the first time unveiled how TFIIH is initially loaded on a carcinogen-adduct DNA lesion by the Rad4 lesion recognition factor, and poised for the subsequent steps of NER including further unwinding of the DNA and lesion verification. Our model agrees with previous biochemical studies and a more recent cryo-EM structure of human TFIIH-XPC. Our study opens the way for structural and mechanistic studies of the entire NER machinery involving all NER components. Structural and definitive biochemical analyses of the downstream NER process is in progress.