Tribbles
Tribology
Tribbles was first identified in Drosophila, where it acts as an important cell cycle regulator. Tribbles proteins function as scaffolding molecules that facilitate the degradation of proteins via a proteasome-dependent mechanism. In Drosophila, tribbles mediates the degradation of the cell division cycle 25 homolog A (CDC25a) homologue string, which regulates cell cycle progression and the CCAAT/enhancer-binding protein (C/EBP) homologue slow border cells (slbo) which functions in oocyte migration. The three mammalian homologs are characterized by a conserved pseudokinase domain and a binding site for the E3 ubiquitin ligase, COP1. Trib proteins lack critical residues for kinase function, and instead, we think that these proteins act as scaffolding molecules to facilitate protein-protein interactions. For more information, check out our review on Trib proteins in normal and malignant hematopoiesis.

Fun fact: Tribbles is named after a Star Trek character. In the 1967 episode "The Trouble with Tribbles", a crew member is gifted an adorable, fluffy Tribble. However, once aboard the Starship Enterprise, the Tribbles begin to proliferate uncontrollably, and threaten to overcome the ship.
The Pear lab first became interested in Trib2, which was identified as a downregulated transcript in leukemic cells undergoing growth arrest. To investigate the effects of Trib2 in hematopoietic progenitors, mice were reconstituted with hematopoietic stem cells retrovirally expressing Trib2, leading to the development of acute myelogenous leukemia. In mechanistic studies, we found that Trib2 associated with and inhibited C/EBPalpha. Furthermore, Trib2 expression was elevated in a subset of human AML patient samples. Read the full article. Subsequent work from the lab showed that, while all three Trib proteins have a conserved binding site, only mice transplanted with hematopoietic progenitors expressing Trib1 and Trib2 (but not Trib3) develop AML. Read the full article.
The expression of the Trib genes in mammals is widely divergent, suggesting the compartmentalization of their function. In resting cells, Trib1 is predominantly expressed in myeloid lineage cells, while Trib2 is generally expressed in lymphoid cells. Trib expression is also altered upon activation in some cell types.
Our main goal is to understand the importance of Trib proteins in immune cell development and function.
Trib proteins as regulators of T cell function
Our recent work (Read the full article here) identified Trib1 as a central regulator of the T cell response to chronic infection. During chronic infection, the magnitude of 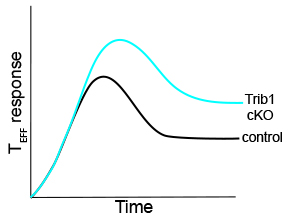 the antiviral T cell response is determined by the balance between T cell function and T cell exhaustion. In chronic disease, persistent antigen stimulation results in a gradual loss in the function and frequency of the exhausted T cell (TEX) pool, and a subsequent loss of pathogen control. Loss of Trib1 enforced a terminal effector cell phenotype within the exhausted setting of chronic infection, enhanced and sustained effector-associated T cell frequency and function, and reduced viral load. These results identify Trib1 as a potential target for enhancing and maintaining durable T cell responses during chronic disease, including cancer. These data support a model of negative feedback regulation whereby Trib1 restrains TCR signaling and downstream functions. As shown in the figure here, we hypothesize that the T cell effector response is enhanced in the absence of Trib1.
the antiviral T cell response is determined by the balance between T cell function and T cell exhaustion. In chronic disease, persistent antigen stimulation results in a gradual loss in the function and frequency of the exhausted T cell (TEX) pool, and a subsequent loss of pathogen control. Loss of Trib1 enforced a terminal effector cell phenotype within the exhausted setting of chronic infection, enhanced and sustained effector-associated T cell frequency and function, and reduced viral load. These results identify Trib1 as a potential target for enhancing and maintaining durable T cell responses during chronic disease, including cancer. These data support a model of negative feedback regulation whereby Trib1 restrains TCR signaling and downstream functions. As shown in the figure here, we hypothesize that the T cell effector response is enhanced in the absence of Trib1.
These data were confirmed and extended by our single-cell sequencing studies that revealed a unique population that was preferentially expanded Trib1 knockout T cells following chronic infection. Although these T cells appeared to be related to exhausted T cells, they were quite different in that the expression of inhibitory receptors was decreased and T cell effector molecules were increased, including multiple members of the killer cell lection-like receptor (KLR) family. Thus, Trib1 appears to be an important determinant of a functional choice between terminally exhausted T cells and the KLR-like T cells that are capable of reactivation. Our studies also provided insights into how Trib1 functions to influence activation versus exhaustion. Rather than promoting degradation, Trib1 binds the protein MALT1 and sequesters it, which ultimately prevents activation downstream of the T cell receptor via the CARMA1-BCL10-MALT1 (CBM) complex, which helps transduce signals from the TCR to the nucleus of T cells.
Future work includes
- Investigating methods to further boost and sustain the effector function of T cells lacking Trib1 during chronic infections and cancer.
- Investigating the immune mechanisms by which Trib1 suppresses T cell function.
- Defining the specific molecular mechanisms by which Trib1 regulates T cell function.
- Developing methods to modulate Trib1 functions as potential therapeutics in chronic infections, autoimmunity, and cancer.
Trib proteins as regulators of immune cell development
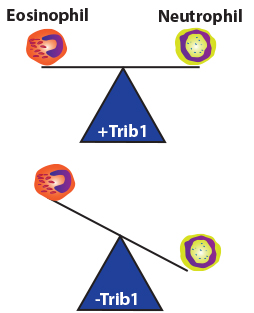 Recent work from our lab identified Trib1 as a key regulator of granulocyte development. Using hematopoietic- and eosinophil-lineage-specific Trib1 deletion, we found that Trib1 regulates both granulocyte precursor lineage commitment and mature eosinophil identity. Conditional Trib1 deletion in hematopoietic stem cells reduced the size of the eosinophil progenitor pool and increased neutrophils, whereas deletion following eosinophil lineage commitment blunted the decrease in eosinophil progenitor pool without increasing neutrophils. These findings provide new insights into early events in myelopoiesis, whereby Trib1 functions at two distinct stages to guide eosinophil lineage commitment and suppress the neutrophil program, promoting eosinophil terminal identity and maintaining lineage fidelity.
Recent work from our lab identified Trib1 as a key regulator of granulocyte development. Using hematopoietic- and eosinophil-lineage-specific Trib1 deletion, we found that Trib1 regulates both granulocyte precursor lineage commitment and mature eosinophil identity. Conditional Trib1 deletion in hematopoietic stem cells reduced the size of the eosinophil progenitor pool and increased neutrophils, whereas deletion following eosinophil lineage commitment blunted the decrease in eosinophil progenitor pool without increasing neutrophils. These findings provide new insights into early events in myelopoiesis, whereby Trib1 functions at two distinct stages to guide eosinophil lineage commitment and suppress the neutrophil program, promoting eosinophil terminal identity and maintaining lineage fidelity.
Future work includes
- Uncovering how Trib1 acts within myeloid progenitors to regulate granulocyte lineage fidelity.
- Identify and characterize how Trib1 functions in neutrophil development and function.
- Understanding the function of Trib1 in specific macrophage functions

