SPACE Study
Serum Proteomic Analysis of Cytokine-Driven Entities (SPACE)
Understanding Cytokine Storms and Inflammatory Disorders
The Serum Proteomic Analysis of Cytokine-Driven Entities (SPACE) study is an international collaboration aimed at understanding the underlying causes of cytokine-driven inflammatory disorders. Cytokine storms—dangerous overreactions of the immune system—can lead to severe illness and death, yet many of these conditions remain poorly understood. Through SPACE, we aim to identify new biomarkers (biological indicators of disease) and potential treatments to improve care for these patients. The study is currently looking for collaborators to provide 40 serum samples from various inflammatory diseases and 5 healthy controls. Using advanced SOMAscan technology, SPACE will analyze over 7,000 protein markers in these samples to uncover potential diagnostic biomarkers and treatment targets for cytokine-driven diseases.
Why It Matters
Cytokine storms are responsible for life-threatening complications in a wide range of diseases, including auto-inflammatory conditions like Castleman disease, Kawasaki disease, and autoimmune diseases like lupus and Crohn's disease, as well as pathogen-induced conditions such as COVID-19, sepsis, and influenza. Despite their impact, few effective treatments exist. Our goal is to change this by uncovering the biological processes behind these conditions and finding new ways to treat them.
How We Do It
SPACE uses cutting-edge technology called SOMAscan to study blood samples from patients with cytokine-driven diseases. By analyzing over 7,000 proteins in each sample, we can identify patterns that indicate how diseases behave and how patients respond to treatments. This method allows us to find new diagnostic markers that can help doctors predict, detect, and treat cytokine storms more effectively.
Goals of SPACE:
- Discover new predictive biomarkers and therapeutic targets common across cytokine-driven disorders.
- Understand the underlying biology of cytokine signaling and immune dysregulation.
- Improve treatments for patients experiencing cytokine storms, by better predicting who will develop them and how they will respond to therapies.
SPACE will tackle critical questions such as:
- Which biomarkers can predict a cytokine storm before it happens?
- What indicators can help doctors choose the best therapy once a cytokine storm begins?
- How can we balance beneficial inflammation with harmful hyperinflammation?
- What long-term effects do treatments have on diseases driven by cytokines?
Using SOMAscan technology, CSTL has already:
- Discovered the first new drug target for iMCD in 25 years (Fajgenbaum et al, JCI, 2019), leading to successful treatment with mTOR inhibitors (Arenas et al, Blood, 2020).
- Identified a second target, leading to the use of JAK1/2 inhibitors for iMCD (Chen et al, Lancet, 2021).
- Discovery that an inflammatory protein called CXCL13, which can be measured in iMCD patients shortly after receiving treatment to help predict whether it will work or not. (Pierson et al, Nature Communications, 2022).
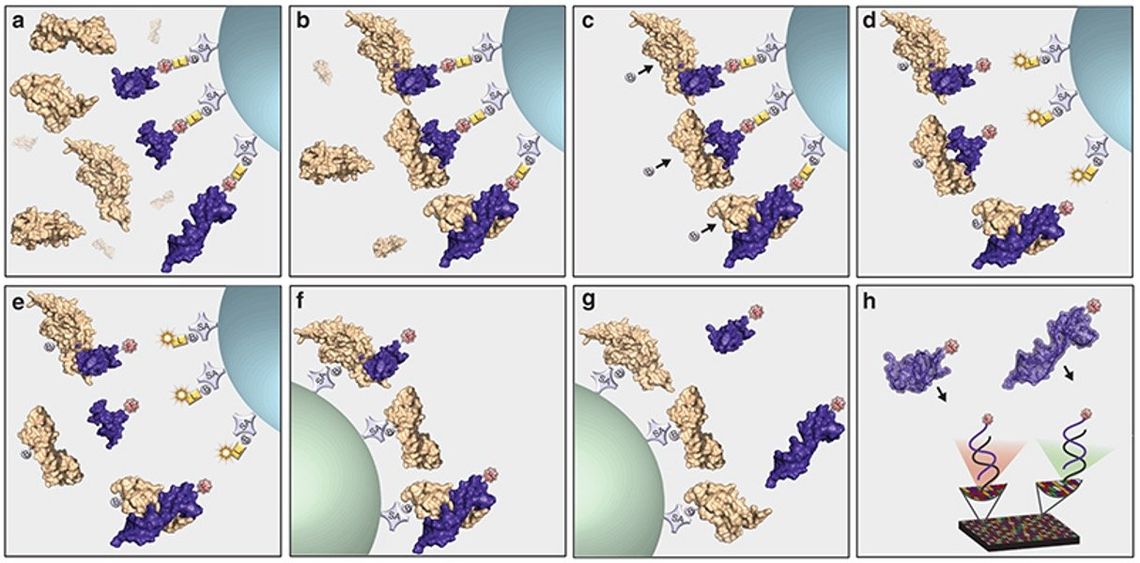
Multiplexed SOMAmer affinity assay. (A) SOMAmer reagents labeled with a 5’ fluorophore, photocleavable linker, and biotin are immobilized on streptavidin (SA)-coated beads and incubated with samples containing a complex mixture of proteins. (B) Cognate (top and bottom) and noncognate (middle) SOMAmer–target protein complexes form on the beads. (C) The beads are washed, removing the unbound proteins and the proteins are tagged with biotin. (D) SOMAmer–protein complexes are released from the beads by photocleavage of the linker with UV light. (E) Incubation in a buffer containing a polyanionic competitor selectively disrupts nonspecific interactions. (F) SOMAmer–protein complexes are recaptured on a second set of streptavidincoated beads through biotin-tagged proteins followed by additional washing steps that facilitate further removal of nonspecifically bound SOMAmer reagents. (G) SOMAmer reagents are released from the beads in a denaturing buffer. (H) SOMAmer reagents are hybridized to complementary sequences on a microarray chip and quantified by fluorescence. Fluorescence intensity is related to protein amount in the original sample. (Adapted from Rohloff et al., 2014.)
Samples needed for SPACE:
| Disease | Priority | Disease Sample Count | Add Healthy Count | Total Count |
| iMCD | 1 | 60 | 5 | 65 |
| HLH (malig.) | 1 | 20 | 5 | 25 |
| HLH (infec.) | 1 | 20 | 5 | 25 |
| HLH (auto.) | 1 | 20 | 5 | 25 |
| IgG4RD | 1 | 40 | 5 | 45 |
| MIS-C | 1 | 40 | 5 | 45 |
| Undiagnosed Inflamm. (includes SURF) | 1 | 40 | 5 | 45 |
| SJIA | 1 | 40 | 5 | 45 |
| SLE | 1 | 40 | 5 | 45 |
| POEMS | 1 | 40 | 5 | 45 |
| HHV8-MCD | 1 | 40 | 5 | 45 |
| Acute COVID | 1 | 40 | 5 | 45 |
| Sepsis | 1 | 40 | 5 | 45 |
| Sjögren's Syndrome | 1 | 40 | 5 | 45 |
| ALPS | 1 | 40 | 5 | 45 |
| Kawasaki Disease | 1 | 40 | 5 | 45 |
| MPS I | 1 | 22 | 5 | 27 |
| VEXAS | 1 | 40 | 5 | 45 |
| Multisystem HR LCH | 2 | 40 | 5 | 45 |
| Erdheim-Chester | 2 | 40 | 5 | 45 |
| Rosai-Dorfman | 2 | 40 | 5 | 45 |
| ANCA-Assoc. Vasculitis | 2 | 40 | 5 | 45 |
| Haploinsuff. of A20 | 2 | 40 | 5 | 45 |
| Rheumatoid Arthritis | 3 | 40 | 5 | 45 |
| Crohn’s/UC | 3 | 40 | 5 | 45 |
| Acute EBV | 3 | 40 | 5 | 45 |
| Acute Influenza | 3 | 40 | 5 | 45 |
| Acute HIV | 3 | 40 | 5 | 45 |
| Dengue Hemorrhagic Fever | 3 | 40 | 5 | 45 |
| CART-induced CRS | 3 | 40 | 5 | 45 |
| Myeloid Neoplasm | 3 | 40 | 5 | 45 |
| AITL | 3 | 40 | 5 | 45 |
| Hodgkin Lymphoma | 3 | 40 | 5 | 45 |
| DLBCL | 3 | 40 | 5 | 45 |
| Indolent Follicular Lymphoma | 4 | 40 | 5 | 45 |
| Long Covid | 4 | 40 | 5 | 45 |
| Multiple Sclerosis | 4 | 40 | 5 | 45 |
| Post-COVID Vaccine | 4 | 40 | 5 | 45 |
| Scleroderma | 4 | 40 | 5 | 45 |
*The table is organized into priority tiers, with Priority Tier 1 representing the highest priority samples and Priority Tier 4 representing the lowest priority.
If you would like to collaborate with us on this project and/or have any questions please reach out to Bridget Austin at bridget.austin@pennmedicine.upenn.edu.


