Cancer Genetics
Cancer Multi-Omics
The post-TCGA (The Cancer Genome Atlas) landscape for high-grade serous ovarian carcinoma (HGSC) reveals a striking genomic complexity. Unlike many cancers, HGSC harbors relatively few recurrent somatic mutations. Instead, it is defined by widespread copy number alterations, creating a genomic terrain of unparalleled intricacy. This complexity underscores the challenge of identifying which alterations are critical drivers of tumorigenesis, tumor viability, and chemotherapy resistance.
Our lab is addressing these challenges by leveraging next-generation sequencing techniques, including whole-exome sequencing (WES), RNA sequencing (RNA-Seq), and ATAC-Seq. These approaches are being deployed to comprehensively characterize the 'omic' alterations in early precursors of HGSC, with a particular focus on BRCA1/2-mutant and CCNE1-amplified tumors. Our work integrates these findings with in vitro and in vivo platforms to uncover vulnerabilities that could inform new therapeutic interventions.
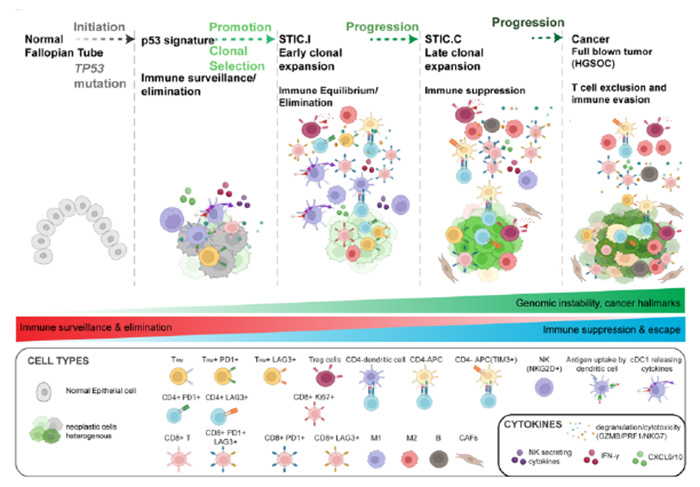
We recently developed the first-ever pre-cancer atlas for HGSC, utilizing high-plex imaging and spatial transcriptomics to map human tissue samples across the spectrum of tumor development. Our findings revealed critical molecular transitions within Fallopian tube precursors, including p53 signatures, serous tubal intraepithelial carcinomas (STIC), and invasive HGSOC. These transitions are characterized by chromosomal instability, persistent interferon (IFN) signaling, and dysregulated immune pathways. We identified early immune modulating mechanisms, such as elevated expression of MHC-class I molecules, including HLA-E, and IFN-stimulated genes, alongside a progressive shift in the tumor microenvironment from immune surveillance in early STICs to immune suppression in advanced lesions and cancer.
While these findings have illuminated new aspects of HGSOC progression, significant questions remain unanswered. How do these immune and genomic alterations converge to drive the transition from precancer to invasive disease? What mechanisms can be leveraged to interrupt this trajectory or restore immune surveillance? The next frontier lies in integrating multi-omic data with emerging technologies such as single-cell profiling, CRISPR-based screens, and advanced imaging to unravel these complexities.

