Research
Research Areas
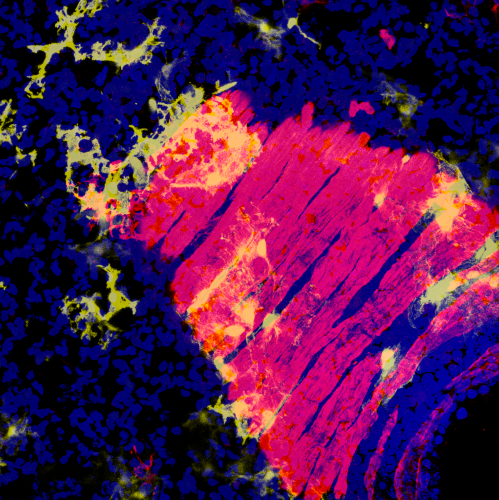 The mature mammalian lung is a highly complex structure comprised of a myriad of different cell types. Our lab has made significant contributions toward understanding the signaling, transcriptional, and epigenetic responses that direct cell fate decisions and structural organization of the lung during embryonic and postnatal development. Understanding these processes provides insight into the developmental defects, both genetic and environmental, that can lead to important pediatric lung diseases and reduce the ability of the immature lung to regenerate in response to injury.
The mature mammalian lung is a highly complex structure comprised of a myriad of different cell types. Our lab has made significant contributions toward understanding the signaling, transcriptional, and epigenetic responses that direct cell fate decisions and structural organization of the lung during embryonic and postnatal development. Understanding these processes provides insight into the developmental defects, both genetic and environmental, that can lead to important pediatric lung diseases and reduce the ability of the immature lung to regenerate in response to injury.
Recent Publications
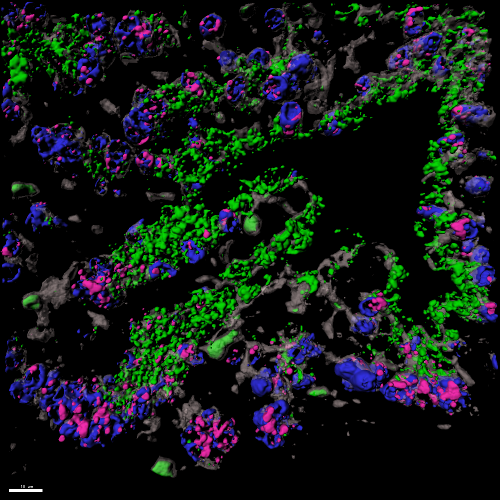 The lung is one of few organs tasked with managing daily environmental insult and injury due to allergens, air pollution, and infectious agents. While the trachea and lungs are relatively quiescent during normal life, they exhibit significant regenerative capacity after acute injury. Our lab employs a variety of models to study both acute and chronic injury to the lung and trachea including animal models, primary human tissue and organoids, and human stem cell models. We are using these approaches to translate basic research findings to better understand the underlying causes of both adult and pediatric lung diseases including Chronic Obstructive Pulmonary Disease (COPD), Pulmonary Hypertension, Idiopathic Pulmonary Fibrosis (IPF), and Congenital Pulmonary Airway Malformations (CPAMs).
The lung is one of few organs tasked with managing daily environmental insult and injury due to allergens, air pollution, and infectious agents. While the trachea and lungs are relatively quiescent during normal life, they exhibit significant regenerative capacity after acute injury. Our lab employs a variety of models to study both acute and chronic injury to the lung and trachea including animal models, primary human tissue and organoids, and human stem cell models. We are using these approaches to translate basic research findings to better understand the underlying causes of both adult and pediatric lung diseases including Chronic Obstructive Pulmonary Disease (COPD), Pulmonary Hypertension, Idiopathic Pulmonary Fibrosis (IPF), and Congenital Pulmonary Airway Malformations (CPAMs).
Recent Publications
Bioinformatics
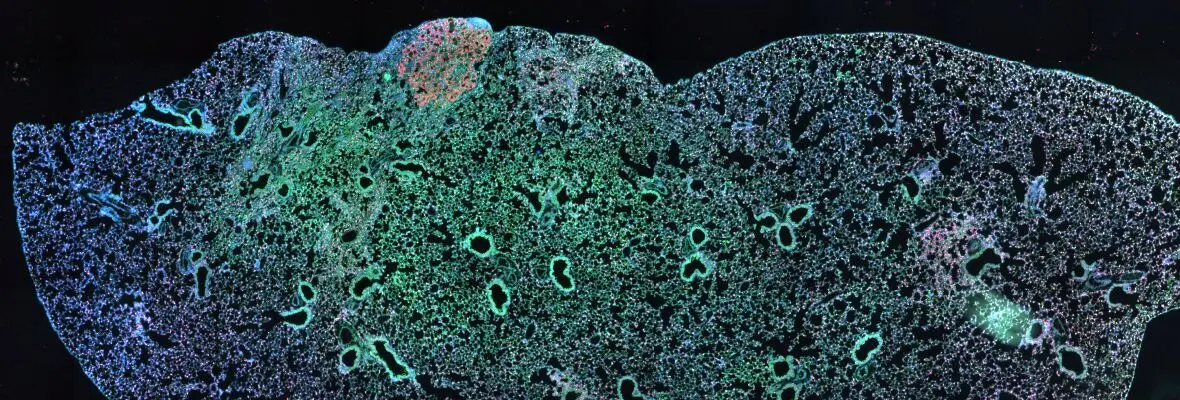
The Morrisey Laboratory has a well-trained bioinformatics team led by Mike Morley. The team works closely with lab members in the design and analysis of genomics experiments. Our Bioinformatics Team has developed cutting edge platforms for lab members and collaborators to visualize and interact with bioinformatics data sets in an iterative manner that allows for rapid rigorous data analysis. Their expertise and tools have allowed for the development of high resolution transcriptomic and epigenetic datasets, both at the cell population and single-cell level, in multiple models and settings including mouse and human, developmental, and the response to regeneration. These data and analyses help to accelerate research discoveries and promote innovation by all lab members.


.png)