Research
Our process identifies scientific challenges in human imaging and pathology and then develops useful solutions through our expertise in the laboratory and collaborations with investigators at the University of Pennsylvania and beyond.
How can we monitor immune, cell, and gene therapies in vivo?
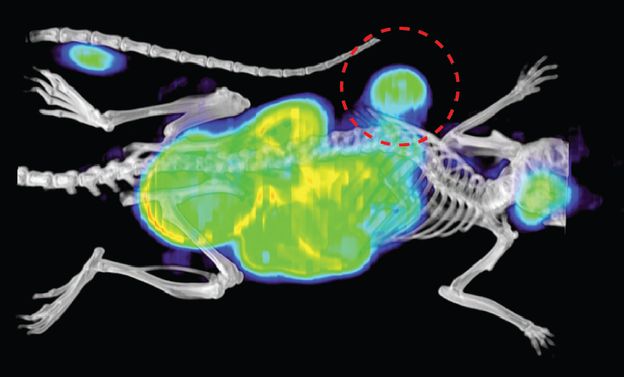
One of the major challenges for immune and cell-based therapies is determining whether the immune system is reaching sites of disease and exerting the desired biological effect. Additionally, imaging tools that can identify immune-mediated toxicities could aid in the evaluation of new therapies or be used clinically to identify toxicities before they become significant or even fatal. We have developed tools that allow longitudinal monitoring of genetically engineered cells, such as CAR-T cells. Our approach uses the sensitivity of positron emission tomography (PET) and repurposes a reporter gene paired with new small molecule probes. The PET imaging pair is based on the high-affinity interaction between bacterial DHFR and the small molecule antibiotic trimethoprim. We are also interested in developing methods that allow assessment of immune system activation at sites of pathology, providing insight into whether therapies are inciting the desired immunologic response.
Related publications:
1. "Monitoring mRNA vaccine antigen expression in vivo using PET/CT." Blizard, G.S., Dwivedi, G., Pan, Y., Hou, C., Etersque, J.M., Said, H., Chevrier, A., Lavertu, M., Ni, H., Davis, B., Tam, Y., Cao, Q., Mach, R.H., Weissman, D., Alameh, M., Sellmyer, M.A. Nat. Commun., 2025, 16 (2234).
2. "Using CD69 PET Imaging to Monitor Immunotherapy-Induced Immune Activation." Edwards, K.J., Chang, B., Babazada, H., Lohith, K., Park, D.H., Farwell, M.D., Sellmyer, M.A. Cancer Immunol. Res., 2022, 10 (9), 1084-1094.
3. "Imaging CAR T Cell Trafficking with eDHFR as a PET Reporter Gene", Sellmyer, M.A., Richman, S.A., Lohith, K., Hou, C., Weng, C.-C., Mach, R.H., O’Connor, R.S., Milone, M.C., & Farwell, M.D. Molecular Therapy, 2020, 28 (1), 42–51.
4. “Quantitative PET reporter gene imaging with [11C]trimethoprim.” Sellmyer, M.A.,* Lee, I., Hou, C., Lieberman, B.P., Zeng, C., Mankoff, D.A., Mach, R.H.* Molecular Therapy, 2017, 25, 120–126. *Co-corresponding authorship.
How can we control the abundance of engineered proteins?
Temporal control of proteins in cells and living animals is crucial to improving the understanding of protein function in the post-genomic era. In addition, technologies that offer control over engineered proteins could be used in therapeutic applications. Since regulation of proteins at a genomic or transcriptional level can be an irreversible or slow process, these tools may not be useful in settings where rapid temporal control is required to achieve immediate knockdown effects. Proteolysis-Targeting Chimeras (PROTACs) have emerged as a strategy to achieve rapid, post-translational control of protein abundance via recruitment of an E3 ligase to the target protein of interest. Our lab has developed several PROTAC molecules by covalently linking the antibiotic trimethoprim (TMP) to pomalidomide, a small molecule ligand of the E3 ligase Cereblon. These molecules induce degradation of various proteins of interest (POIs) genetically fused to E. coli dihydrofolate reductase (eDHFR), the molecular target of TMP. We demonstrate that various eDHFR-tagged proteins can be knocked-down in vitro and in preliminary mouse models. TMP PROTACs are a powerful addition to the suite of TMP-eDHFR tools that can be used in concert with TMP diagnostic radiotracers as a “theranostic” pair. We aim to further our PROTAC technology to achieve modulation of more advanced cellular functions as well as multiplexed protein regulation within single cells.
Related publications:
- "Regulation of eDHFR-tagged proteins with trimethoprim PROTACs." Etersque, J.M., Lee, I.K., Sharma, N., Xu, K., Ruff, A., Northrup, J.D., Sarkar, S., Nguyen, T., Lauman, R., Burslem, G.M., Sellmyer, M.A. Nat. Commun., 2023, 14 (7071).
- "A genetically encoded protein tag for control and imaging of CAR T cell therapy." Lee, I.K., Sharma N., Noguera-Ortega, E., Liousia, M., Baroja, M.L., Etersque, J.M., Pham, J., Sarkar, S., Carreno, B.M., Linette, G.P., Pure, E., Albelda, S.M., Sellmyer, M.A. Mol. Ther., 2023, 31 (12), 3564-3578.
How can we separate pathologies with overlapping characteristics, including inflammation, infection, and cancer?
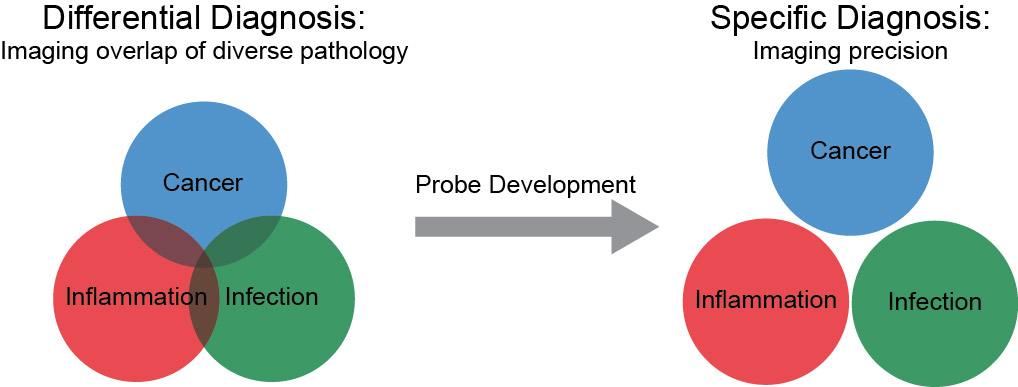
These projects are rooted in a common clinical scenario where a diagnosis is not straightforward. Physicians do not always know whether a patient’s particular symptom, lab value, or imaging finding is due to inflammation, infection, or cancer. As there is overlap in both clinical and imaging findings, a specific imaging agent that could distinguish bacterial infection from these other pathologies would be of great utility. Moreover, such an agent could allow monitoring of treatment of challenging bacterial infections (i.e. in the setting of prior surgery or cancer treatments). Using a well-known antibiotic, trimethoprim (TMP), we developed carbon-11 and fluorine-18 labeled positron emission tomography (PET) radiotracers. Contrary to trends in the field which have shifted to using metabolic approaches to imaging bacteria, we have shown that these antibiotic-based radiotracers can be used to image bacterial infection in rodent models. Importantly, radiolabeled trimethoprim did not show confounding uptake in sterile inflammation or cancer, etiologies that limit the specificity of other long-standing nuclear technologies. We are currently testing the breadth of organisms that can be identified with these radiotracers, and we have an active clinical protocol in human patients with suspected bacterial soft tissue and lung infections. These tools could be important for our understanding the host-pathogen interaction especially in patients with chronic infections and with underlying pre-disposing diseases such as cystic fibrosis and diabetes.
Related publications:
1. "Imaging sensitive and drug-resistant infection with [11C]-trimethoprim." Lee, I.K.*, Jacome, D.A.*, Cho, J.K., Tu, V., Young, A.J., Dominguez, T., Northrup, J.D., Etersque, J.M., Lee, H.S., Ruff, A., Aklilu, O., Bittinger, K., Glaser, L.J., Dorgan, D., Hadjiliadis, D., Kohli, R.M., Mach, R.H., Mankoff, D.A., Doot, R.K., Sellmyer, M.A. J. Clin. Invest., 2022, 132 (18), e156679. *Co-first authorship.
See Highlights: "Imaging infection and inflammation." J. Clin. Invest., 2022, 132 (18), e162885.
2. "Bacterial infection imaging with [18F]fluoropropyl-trimethoprim." Sellmyer, M.A., Lee, I., Hou, C., Weng, C.-C., Li, S., Lieberman, B.P., Zeng, C., Mankoff, D.A., Mach, R.H. Proc. Natl. Acad. Sci., 2017 114(31): 8372-8377. *Co-corresponding authorship.
See Highlights: Spotlight. Chem. Res. Toxicol. 30, 1653–1654 (2017).
How can we modulate and sense cell-cell interactions in live animals?
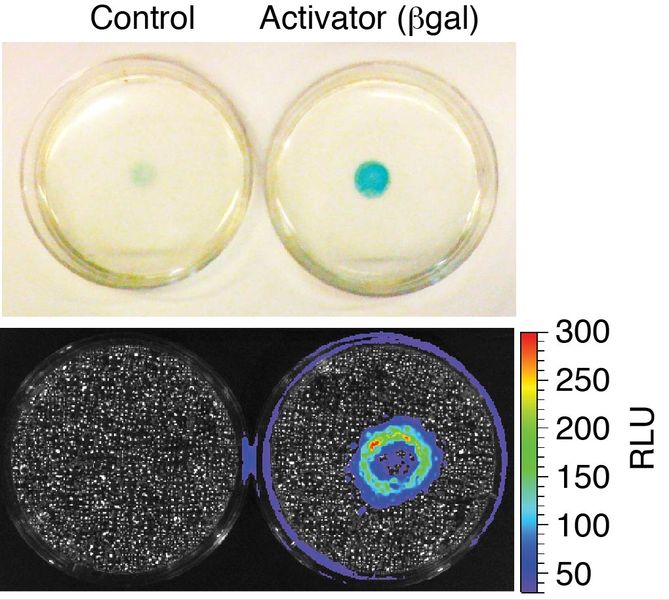
We develop molecular systems using principles of synthetic biology which allow cells to report on the activities of other nearby cells. This approach, termed cell-based diagnostics, is especially exciting the with the recent advances in modern cell-based therapeutics such as CAR-T cells.
For example, we developed a first-in-class technology to report when two cell populations interact or come into close proximity in an animal using bioluminescent imaging. The “proximity reporter” is based on a caged luciferin and sequential enzyme catalysis. In the petri-dish picture above, reporter cells nearest to activator cells that encode beta-galactosidase are exposed to higher local concentrations of luciferin – resulting in a higher bioluminescent signal (hence the ring in the middle of the plate). We used the proximity reporter to find sites of immune surveillance in a metastatic breast cancer model that would have otherwise escaped conventional imaging techniques. Our work was highlighted in Science and Nature Methods, and was an important milestone in developing synthetic biology tools to observe cellular interactions in vivo on a whole animal scale. We are currently working on the next generation tools for imaging the immune system and its relationship to cancer and infection.
Related publications:
1. "A Chemical Approach for Programmable Protein Outputs Based on Engineered Cell Interactions." Jacome, D.A., Northrup, J.D., Ruff, A.J., Reilly, S.W., Lee, I.K., Blizard, G.S., & Sellmyer, M.A. ACS Chemical Biology, 2021, 16 (1), 52-57.
2. “Visualizing Cellular Interactions with a Generalized Proximity Reporter.” Sellmyer, M.A., Bronsart, L., Imoto, H., Contag, C.H., Wandless T.J., Prescher, J.A. Proc. Natl. Acad. Sci., 2013, 110, 8567-8572.


