Projects
Work in our laboratory is aimed toward the understanding of molecular pathways that govern chronic kidney disease development.
1. Patient cohorts and tissue samples.
One key bottleneck to progress in our field has been the lack of well-characterized human tissue samples. To address these issues, we have (a) established the largest human kidney tissue bank (b) launched longitudinal patient cohort studies, where tissue samples, associated non-invasive biofluids are associated with outcome in a prospective manner.
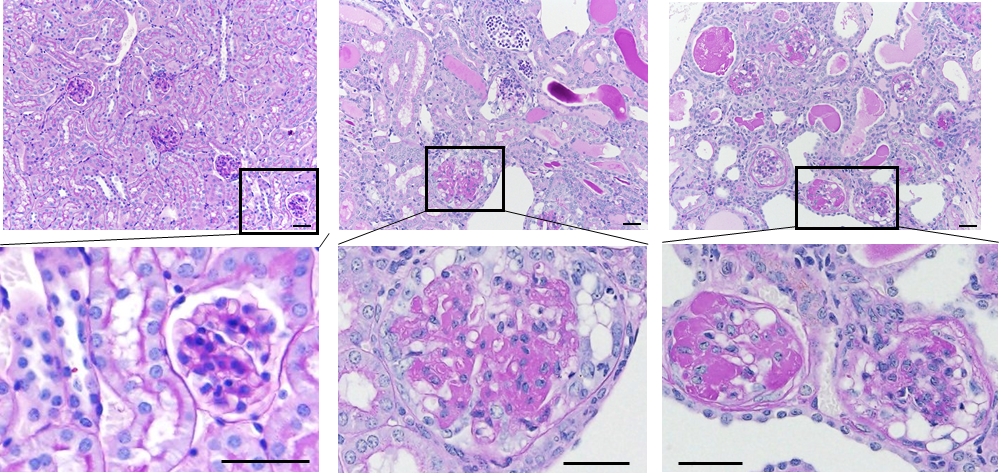
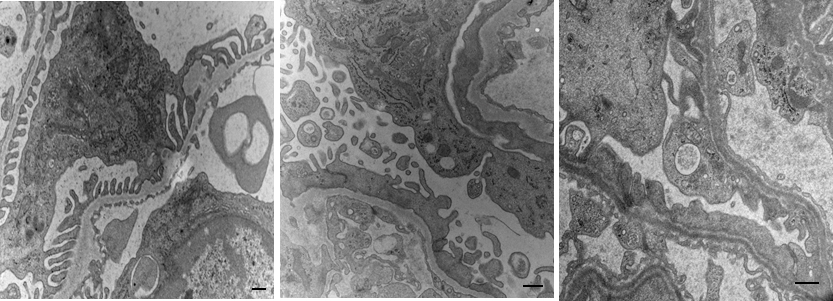
Susztaklab Biobank: We collect human kidney samples from clinically indicated nephrectomies, living donors and clinical biopsy samples (n>1,600).
- Clinical and demographics information is collected and recorded in a database using an “honest broker” system.
- Follow-up clinical information focusing on kidney function and life status.
- Formalin-fixed and paraffin-embedded kidney tissue samples.
- Traditional- and computer-aided histopathological analysis
- Fresh tissue samples, samples stored in RNALater and primary cell lines.
Cohort studies with deep molecular and cellular phenotypes:
- TRIDENT (Transformative Research In Diabetic NEphropaThy) is a multi-center observational cohort that plans to enroll and follow 400 subjects with diabetes. You can read about Trident at http://www.med.upenn.edu/trident/
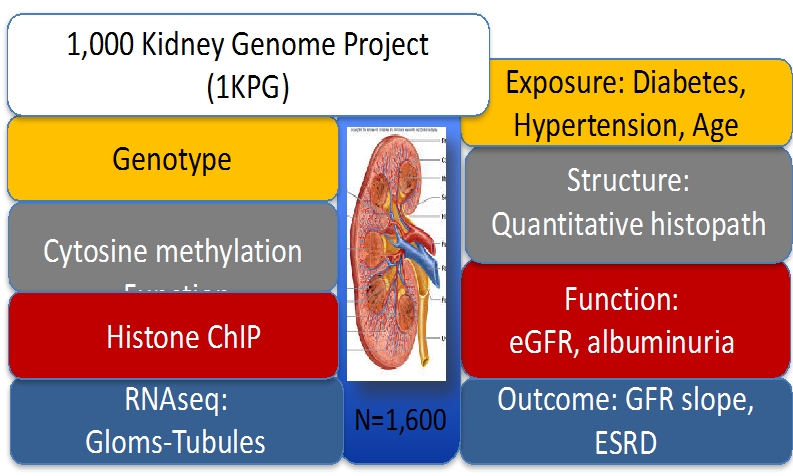
2. 1KGP (1,000 Kidney Genome Project) a comprehensive multi-scalar dataset for the human kidney
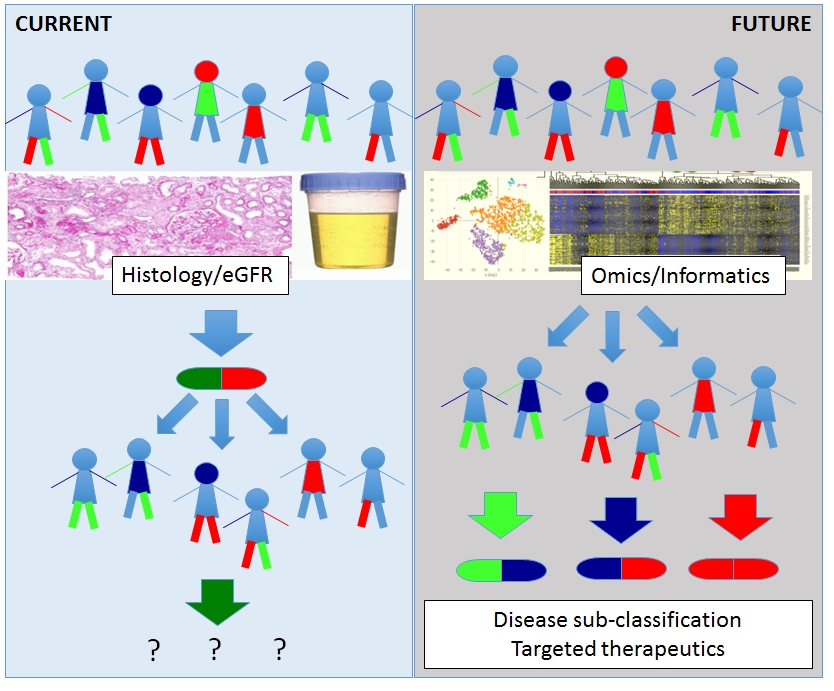
We hypothesize that integrative analysis of epigenetic and genetic settings in healthy and diseased cells can provide a rational basis for more accurately modeling the critical biological pathways involved in mediating the progressive phenotype in individual patients. We also predict that epigenomic integrative analysis can be used to determine the identity of chromatin and transcription factors that contribute mechanistically to aberrant transcriptional programming in chronic kidney disease, and that this information can be used for designing therapeutic strategies . The following datasets are generated to achieve these goals.
- Genotype information
- Genome wide cytosine methylation datasets; Whole Genome Bisulfate sequencing, Methylation BeadChip Arrays (450K, EPIC)
- Epigenome data (histone ChiP)
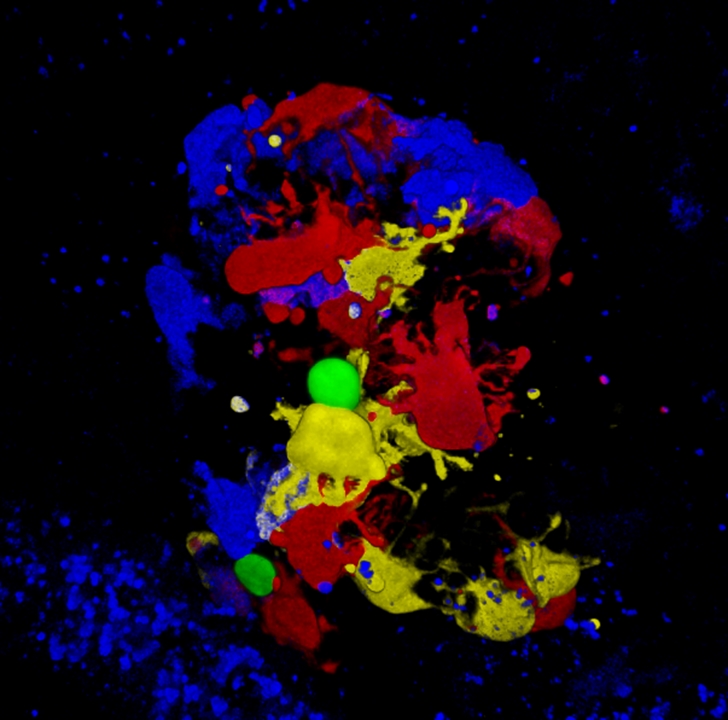
- Tissue and compartment specific (glomerular and tubule) RNAsequencing
- Transcriptome and epigenome analysis at the single cell level
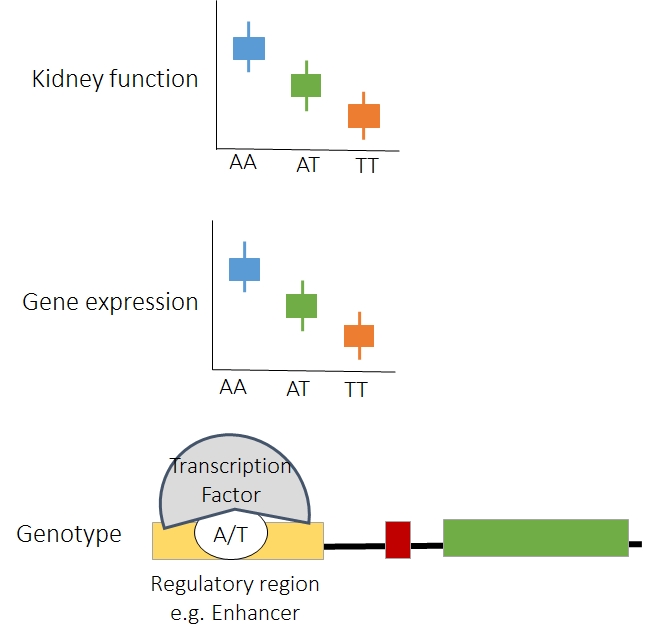
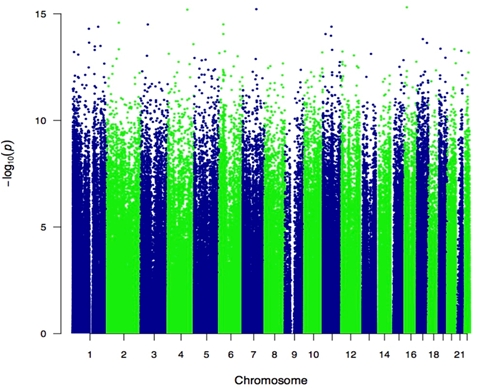
3. Understanding the role of genetic variation in renal phenotypic variation.
Major focus of our lab is to understand how genetic variations lead to phenotype development. Particularly we are interested in understanding the association between genotype driven gene expression and cellular phenotype variation, particularly focusing on genetic variants that are known to be enriched in patients with kidney disease. We detect functional genetic variation in regulatory elements and subsequently use regulatory variation and accurately measured gene expression variation to bridge the genotype with disease phenotypes in association studies. We take advantage of our human kidney tissue bank as we generated compartment-specific expression data (using RNAsequencing) and genotype information. The epigenome datasets further helps to narrow genetic variation that are harbored within functional elements.
4. Defining the role of epigenetic changes in kidney disease development
Epigenome-wide association studies represent one means of applying genome-wide assays to identify molecular events that could be associated with human phenotypes. The epigenome is especially intriguing as a target for study, as epigenetic regulatory processes are, by definition, heritable from parent to daughter cells and are found to have transcriptional regulatory properties. As such, the epigenome is an attractive candidate for mediating long-term responses to cellular stimuli, such as environmental effects modifying disease risk.
5. Understanding kidney disease development by in vivo (mouse) and in vitro (cellular) models.
Traditionally, we used the Cre/loxP and tet inducible transgenic technologies which allowed us to analyze the function of particular factors by deleting or overexpressing genes that encode in specific cell types in the kidney.
Recently, we use the CRIPR-Cas9 system to generate cell-based and animal models for kidney disease. We are pursuing different pathways based on our large scale discovery studies as described below.
- We are working on determining the role of the Notch and Wnt/beta-catenin pathway in chronic kidney disease development, renal epithelial cell homeostasis, renal stem or progenitor cell function and differentiation. Our recent results highlight the role of embryonic programs in adult disease development (Niranjan et al Nature Medicine 2008, Bielesz et al JCI 2010).
- The kidney has very high energy demand. Our recent studies indicate the critical role of metabolism and nuclear transcription factors in kidney disease development (Kang et Nature Medicine 2015).
- We are developing and characterizing novel mouse models for human genetic variation. Such as we developed a model for G1 and G2 APOL1 risk variants (Beckerman and Bi-Karchin Nature Medicine 2017). This model recapitulates features of kidney disease seen in patient with genetic variants.