Projects
FRAMEWORK:
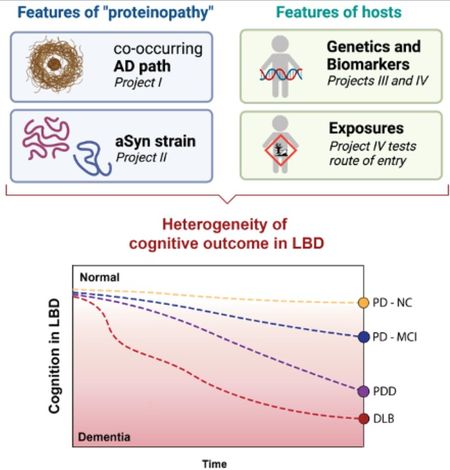
We hypothesize that features of the proteinopathy and features of the host act in concert to determine the pace and pattern of alpha-synuclein spread in the brain. This pace and pattern in turn determines how cognition manifests in the individual patient.
Projects
Project Leader David Irwin
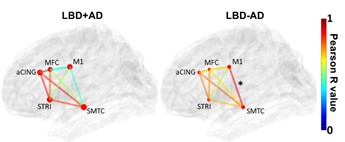
Project I establishes the “ground truth” with respect to the relationship between aSyn pathology and patterns of neuronal connectivity in the human brain while integrating postmortem data with antemortem MRI findings from the same individual. Project I’s first aim stratifies LBD cases into those with vs. without concomitant AD pathology (LBD+AD vs. LBD-AD), then compares networks of aSyn pathology found in LBD+AD vs. LBD-AD in >300 autopsied LBD cases using digital histopathology techniques and graph theory. This novel approach allows for the simultaneous examination of complex molecular interactions across brain regions that are not possible with standard statistical modeling and histopathology techniques. Project I’s second aim drives its postmortem findings forward into the antemortem stage, by analyzing our unique resource of neuropathology cases with longitudinal MRI collected by the Clinical Core during life, identifying structural features on imaging that correspond to pathological findings and clinical heterogeneity of LBD.
Project Leader Virginia Lee
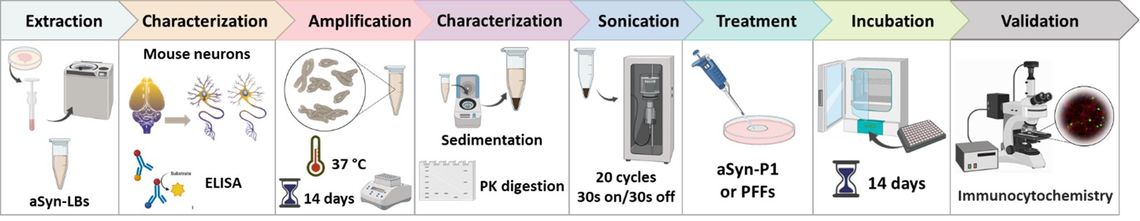
Project II investigates the contribution of aSyn “strain” differences to the observed heterogeneity in cognitive outcomes among individuals with LBD. With notable exceptions, investigation of aSyn strain differences has relied on artificial strains of aSyn pre-formed fibrils (PFFs). However, aSyn PFFs do not fully recapitulate the biological behavior of Lewy bodies, motivating Project II’s goal of characterizing the ultrastructure and cell biological characteristics of human brain-derived aSyn strains. Project II benefits from an important recent advance in the Lee laboratory: the development of techniques to amplify aSyn species from human brain tissue using human brain aSyn as template and monomeric aSyn as substrate. Because amplified aSyn species preserve biochemical and cell biological characteristics of the source material, both the source material and the amplified aSyn (termed LBs-P1) can be evaluated and compared with aSyn PFFs derived from recombinant material. Project II’s first aim will employ cryo-electron tomography (cryo-ET) for ultrastructural analyses of cells treated with human brain-derived aSyn or LBs-P1, alongside expansion microscopy to physically expand cells in order to gain nanoscale resolution in a “de-crowded” cell environment. Project II’s second aim investigates mechanisms of cellular uptake of LBD brain-derived aSyn, following up on preliminary work suggesting that uptake mechanisms may differ between PFFs and aSyn derived from human LBD. Using both fixed and live cell imaging, uptake, transport, and neuronal transfer over time will be studied for multiple potential strains of LBD brain-derived aSyn and LBs-P1, compared with PFFs, in neuronal culture. Project II’s final aim focuses on the effect of LBD brain-derived aSyn on lysosomal function. Importantly, Project II will marry ultrastructural and cell biological characterization, allowing insight into the basis of “strain” differences in aSyn.
Project Leader Alice Chen-Plotkin
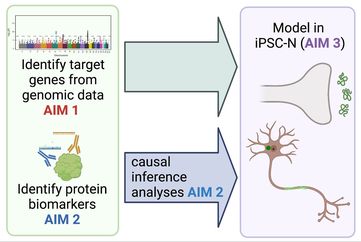
Project III uses the wealth of data represented by the genomewide association studies (GWAS) that have already been performed in tens of thousands of PD and DLB patients. While these GWAS have produced 90+ loci, the target genes to which these loci belong, and the biological functions of these target genes, are largely unexplored. To fully capitalize on the power of these genomic studies, Project III’s first aim will assign target genes to the 90+ loci nominated by GWAS for risk variants associated with PD, risk variants associated with DLB, and risk variants associated with cognitive decline in PD. Project III’s second aim will then screen CSF samples for proteins associating with cognitive outcome in PD using a proximity ligation-based assay platform that can screen >1000 proteins, cross-referencing CSF hits against plasma biomarkers found on our previously-performed “omic”-scale biomarker screens to associate with this phenotype. This aim will also perform Mendelian-randomization-based causal inference analyses for top CSF and plasma biomarkers associating with cognitive outcome in PD, in order to determine whether a given biomarker might be causal for cognitive decline. Project III’s final goal will be to move the most promising 10-20 targets into neuronal model system where they can be manipulated using CRISPR-Cas9 editing, in order to determine the effect of these “omics”-derived leads.
Project Leader Kelvin Luk
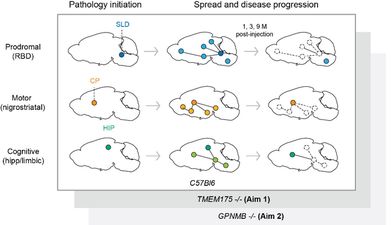
Project IV extends the work of investigating the effects of host genetic background and routes of aSyn exposure to in vivo models. Specifically, we build on our recent success elucidating mechanisms behind the association of TMEM175 and GPNMB with PD risk, performing the detailed mouse studies that are necessary to understand how cellular effects translate into whole-brain or whole-organism effects. Project IV’s first aim extends our prior finding that PD-associated TMEM175 variants decrease function of the TMEM175 lysosomal potassium channel, investigating the pattern of pathological aSyn spread, neurodegeneration, and organismal phenotype in TMEM175 knockout animals following injection of aSyn PFFs to the subcoeruleus (to model prodromal LBD), hippocampus (to model limbic LBD) vs. striatum (baseline condition, modeling motor LBD). In Aim 2, we extend our prior finding that PD-associated GPNMB variants increase GPNMB expression, allowing for more cell-to-cell spread of aSyn pathology, using a similar experimental paradigm as in Aim 1. Here, however, we will inoculate GPNMB knockout animals vs. controls. For both Aims 1 and 2, 1-month, 3-month, and 9-month timepoints will be assessed, with an 18-month timepoint for Aim 2 as well, in order to capture the temporal element of spread of synucleinopathy and its consequences. In Aim 3, human brain-derived aSyn LBs-P1 from Project II will be introduced, in the GPNMB and TMEM175 backgrounds, in order to elucidate the genetic background x aSyn strain interaction in an animal model.

