Projects
Impact and Vision
By integrating human genetics, cell-resolved biology, longitudinal cohorts, and AI-enabled modeling, our work creates a direct path from mapping → mechanism → medicine in kidney disease.
Mapping to Mechanism to Medicine
Our research strategy moves systematically from human genetic discovery, to cellular and molecular mechanism, to translational insight and therapeutic opportunity. We integrate large-scale human data, experimental validation, and AI-enabled modeling to define disease-driving pathways in chronic kidney disease.
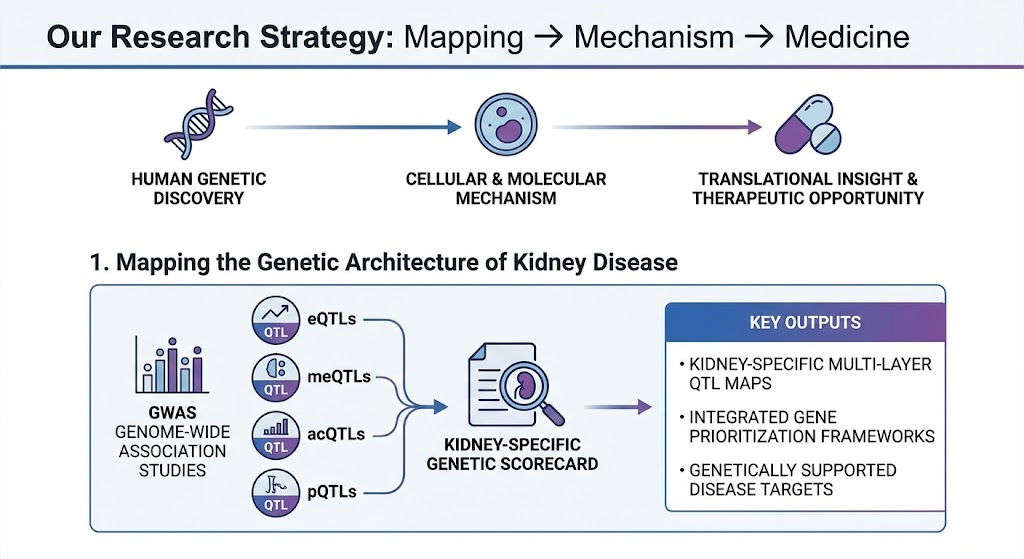
1. Mapping the Genetic Architecture of Kidney Disease
Human genetics provides an unbiased entry point into kidney disease biology.We integrate genome-wide association studies with kidney-specific molecular quantitative trait loci (eQTLs, meQTLs, acQTLs, pQTLs) to identify and prioritize causal genes and regulatory variants.
This work established one of the first kidney-specific genetic scorecards, enabling systematic prioritization of disease genes based on convergent genetic, epigenomic, and transcriptional evidence.
Key outputs
-
Kidney-specific multi-layer QTL maps
-
Integrated gene prioritization frameworks
-
Genetically supported disease targets
Selected publications
-
Park et al., Science (2018)
-
Qiu et al., Nature Medicine (2018)
-
Sheng et al., Nature Genetics (2021)
-
Liu et al., Nature Genetics (2022)
2. Defining Cell-Type–Specific Disease Mechanisms
Genetics alone does not explain the full spectrum of kidney disease.To resolve disease mechanisms, we generated comprehensive human kidney cell atlases using single-cell and spatial transcriptomics.
These studies demonstrated that specific disease phenotypes are driven by dysfunction in defined cell populations, providing a cellular framework for understanding progression, fibrosis, and inflammation.
Key discoveries
-
Injury-associated epithelial cell states
-
Disease-specific immune and stromal niches
-
Spatial organization of pathogenic cell interactions

Selected publications
-
Kang et al., Nature Medicine (2015)
-
Chung et al., Cell Metabolism (2019)
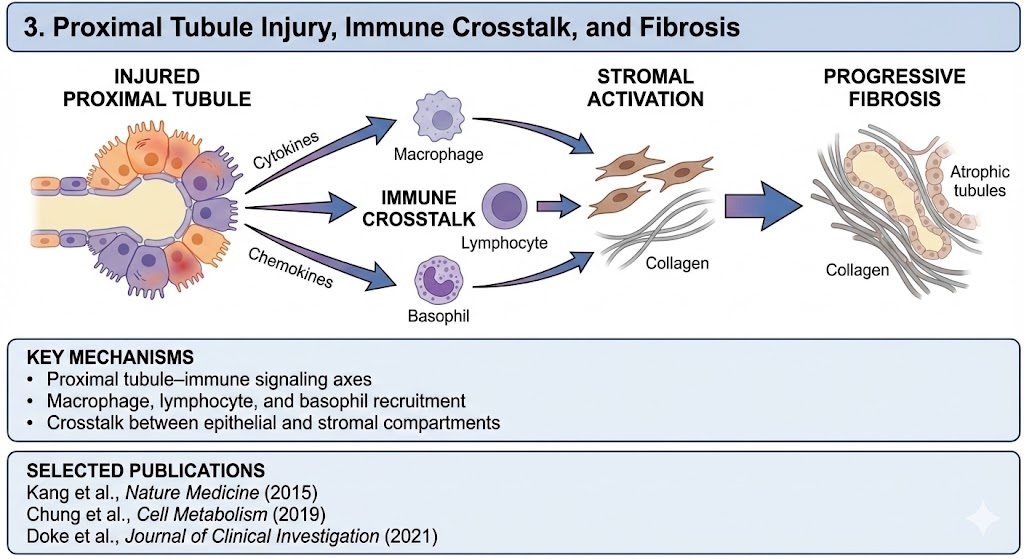
3. Proximal Tubule Injury, Immune Crosstalk, and Fibrosis
A central theme of our work is the role of the injured proximal tubule as an active organizer of kidney inflammation and fibrosis, rather than a passive victim of injury.
We demonstrated that genetically and metabolically injured proximal tubule cells secrete specific cytokines and chemokines that orchestrate immune cell recruitment and stromal activation, driving progressive fibrosis.
Key mechanisms
-
Proximal tubule–immune signaling axes
-
Macrophage, lymphocyte, and basophil recruitment
-
Crosstalk between epithelial and stromal compartments
Selected publications
-
Kang et al., Nature Medicine (2015)
-
Chung et al., Cell Metabolism (2019)
-
Doke et al., Journal of Clinical Investigation (2021)
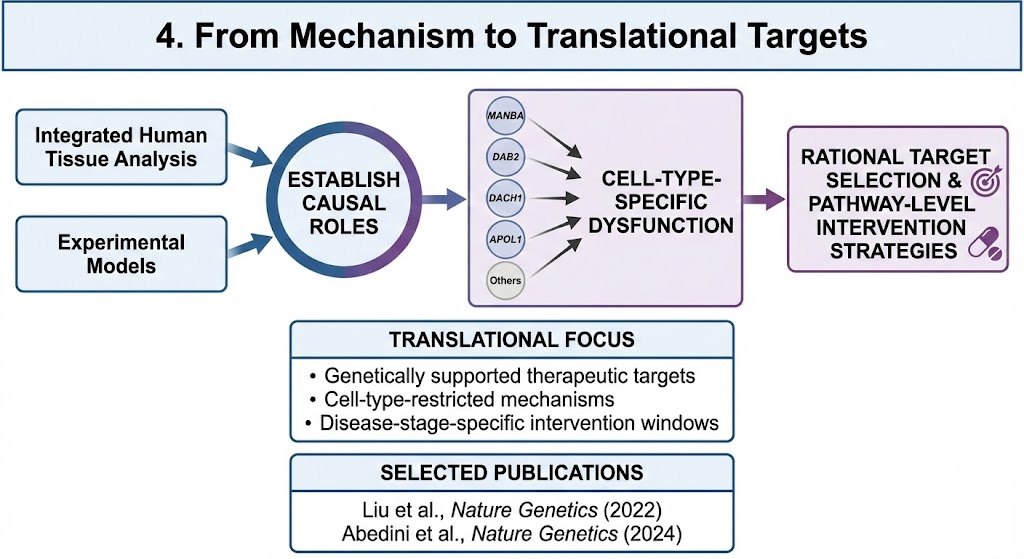
4. From Mechanism to Translational Targets
We use integrated human tissue analysis and experimental models to establish causal roles for genetically supported disease genes, including MANBA, DAB2, DACH1, APOL1, and others.
This work connects genetic risk to cell-type-specific dysfunction, enabling rational target selection and pathway-level intervention strategies.
Translational focus
-
Genetically supported therapeutic targets
-
Cell-type-restricted mechanisms
-
Disease-stage-specific intervention windows
Selected publications
-
Liu et al., Nature Genetics (2022)
-
Abedini et al., Nature Genetics (2024)
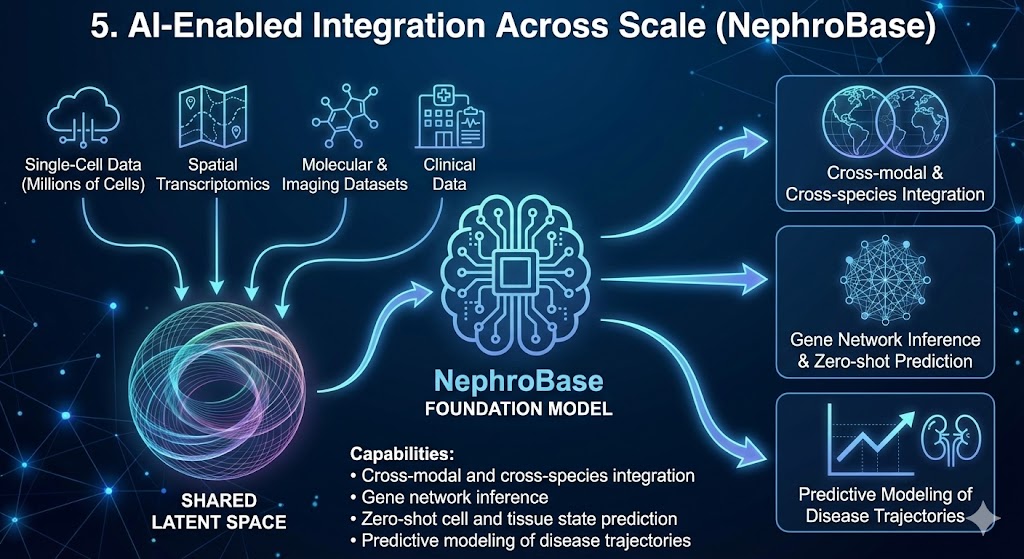
5. AI-Enabled Integration Across Scale (NephroBase)
The scale and complexity of kidney data now exceed what traditional analysis approaches can handle. To address this, we are developing NephroBase, the first kidney-specific foundation model.
NephroBase integrates tens of millions of cells across single-cell, spatial, molecular, imaging, and clinical datasets into a shared latent space, enabling zero-shot prediction, cross-dataset integration, and scalable discovery.
Capabilities
-
Cross-modal and cross-species integration
-
Gene network inference
-
Zero-shot cell and tissue state prediction
-
Predictive modeling of disease trajectories