Learning Health System Research
As an academic medical center focused on learning health system research, Penn Medicine has a variety of data access and access points to support clinical research.

Penn Medicine Self Service Data Options
The Penn Medicine Biobank (PMBB) is a research program at Penn for which any patient may sign up. There is an overarching consent that is presented to patients when they engage in patient activity within Penn Medicine. The PMBB includes storage of DNA, plasma, serum and other selected tissues and can be combined with clinical data via PennChart. The PMBB also provides whom exome sequencing as well as genotyping.
If you would like to request access to data for your protocol please discuss with the PMBB and team and begin the process here. Computing and storage options include on premise high performance computing clusters and cloudbased computing options through DNANexus and Azure.
RADAR is Penn Medicine’s imaging and analytics resource. The RADAR team also maintains up to date lists of imaging equipment and can help support teams with grants or medical imaging data as needed. Requests and more details can be found here.
Institute for Biomedical Informatics (IBI) Clinical Research Informatics Core (CIC) provides data brokering and predictive analytics services of clinical data for clinical and translational research in which the goal is to learn actionable healthcare knowledge and develop impactful solutions for improving patient care.
The Electronic Phenotyping Resource (EPR) is a shared resource of the Abramson Cancer Center dedicated to unlocking the power of electronic health records for clinical and translational research in cancer care.
The EPR provides consultative and technical services that leverage the oncology informatics team's deep expertise in clinical research informatics, intimate knowledge of the clinical information systems and workflows in use throughout the Penn Medicine Cancer Service Line, and unique and powerful "real world" data assets, including:
- The Oncology Research and Quality Improvement Datamart (ORQID) – a data warehouse that aggregates together structured data extracted from PennChart (Penn's instance of the Epic EHR), the Penn Medicine Cancer Registry (accredited by the American College of Surgeons Commission on Cancer), the ARIA® Radiation Oncology Information System, the Penn Center for Personalized Diagnostics genomic variant database, and several other clinical systems
- Disease-specific, human-curated research datasets developed in partnership with Flatiron Health, Inc.
- The ACC Catchment Explorer (ACE) - a self-service data exploration tool which provides access to population-level incidence and mortality statistics across our the ACC catchment area, as well as ACC-specific case volume
Center for Data, Outcomes and Tea Science in GI. CDOTS is a working group of GI investigators that are working together make research data collection easier. The group’s objective is to build a data and regulatory infrastructure to facilitate efforts to gather preliminary research data for grants and attract outside funding. Clinical Data requests or project support can be found here The Center for Data, Outcomes, & Team Science (CDOTS) | Serper Research Group | Perelman School of Medicine at the University of Pennsylvania
Coming Soon
Coming Soon
There are several highly trained individuals who work within departments to pull or wrangle data for researchers. These are designated full time staff members who support clinical research within the various departments. In addition, they are overseen by the Office of Clinical Research and Learning Health system core to ensure compliance with HIPAA and institutional policies. Your request may be triaged to an honest broker based on department, type of research and/ or capacity.
Honest Broker Service Request Intake - Data Request | PennDnA | Perelman School of Medicine at the University of Pennsylvania
Epic SlicerDicer is a query tool that can be helpful in generating cohorts of research subjects in PennChart or eHealth, assessing study feasibility and help to identify subjects for trial recruitment. You need Epic access to use SlicerDicer.
For SlicerDicer training: uphsnet.uphs.upenn.edu/AnalyticsStorefront/Home/education
For SlicerDicer general info: Slicer Dicer | Penn Medicine Clinical Research | Perelman School of Medicine at the University of Pennsylvania
Coming soon – SlicerDicer office hours available via KnowledgeLink sign-up and will be offered once a month.
Epic Cosmos is a data resource made up of data from participating Epic health care systems, including UPHS. It is a deidentified data set and can be accessed with the appropriate training either for the SlicerDicer tool or data scientist tool.
More information on the Epic Cosmos data can be found at: Cosmos | Penn Medicine Clinical Research | Perelman School of Medicine at the University of Pennsylvania
TriNetX is a large research network which includes the PennChart EHR data that is mapped using the OMOP (Observational Medical Outcomes Partnership common data model). The data set is deidentified and the platform contains a number query and statistical summary tools. Penn Medicine data can be isolated and depending upon study needs, you can request a data set from the TriNetX network as well as request linked data to PennChart patients if needed.
For detailed training (and access request) – TriNetX makes available several training resources: TriNetX | Penn Medicine Clinical Research | Perelman School of Medicine at the University of Pennsylvania
PennChart Research Learning Labs are offered once a month and provide ongoing PennChart education for researchers. These hands-on courses range from refreshers on existing tools, to demonstrations of new and upcoming features in PennChart. We regularly update our topics to meet the needs of the Penn research community and to incorporate new features as they become available. These sessions will include hands-on demonstrations, followed by a Q&A session where researchers can seek assistance with specific projects.
Register for an upcoming PennChart Research Learning Lab.
Topics may include:
- Research Documentation in PennChart
- Research Billing in PennChart
- New Features Demo in PennChart
- Informed Consent Module
- Adverse Event Module
- SlicerDicer
The honest broker program is a mechanism for your department or center to have someone trained and certified to directly extract research data from Epic PennChart and eHealth (Clarity and Caboodle databases).
The link below has a brief description of the program and there is an email to OCR where if you have someone interested, they can email for more information and to initiate the process:
Honest broker training requirements:
- CITI human subjects training
- HIPAA training
- Sensitive data restrictions review
- Attestation to follow policies & guidelines
- OCR & DnA approval
To access Epic records as an honest broker:
- Completion of certification, accreditation or proficiency* level via Epic
- Cogito: 3-days, virtual; $1,200; project and exam required
- Caboodle: 1-day, virtual; $400; exam required
- Clarity: 1.5-days, virtual; $600; exam require
*Certification requires in-person classes (Verona, WI); accreditation requires virtual classes; proficiency requires self-study (no fees or class attendance). All require exams / project.
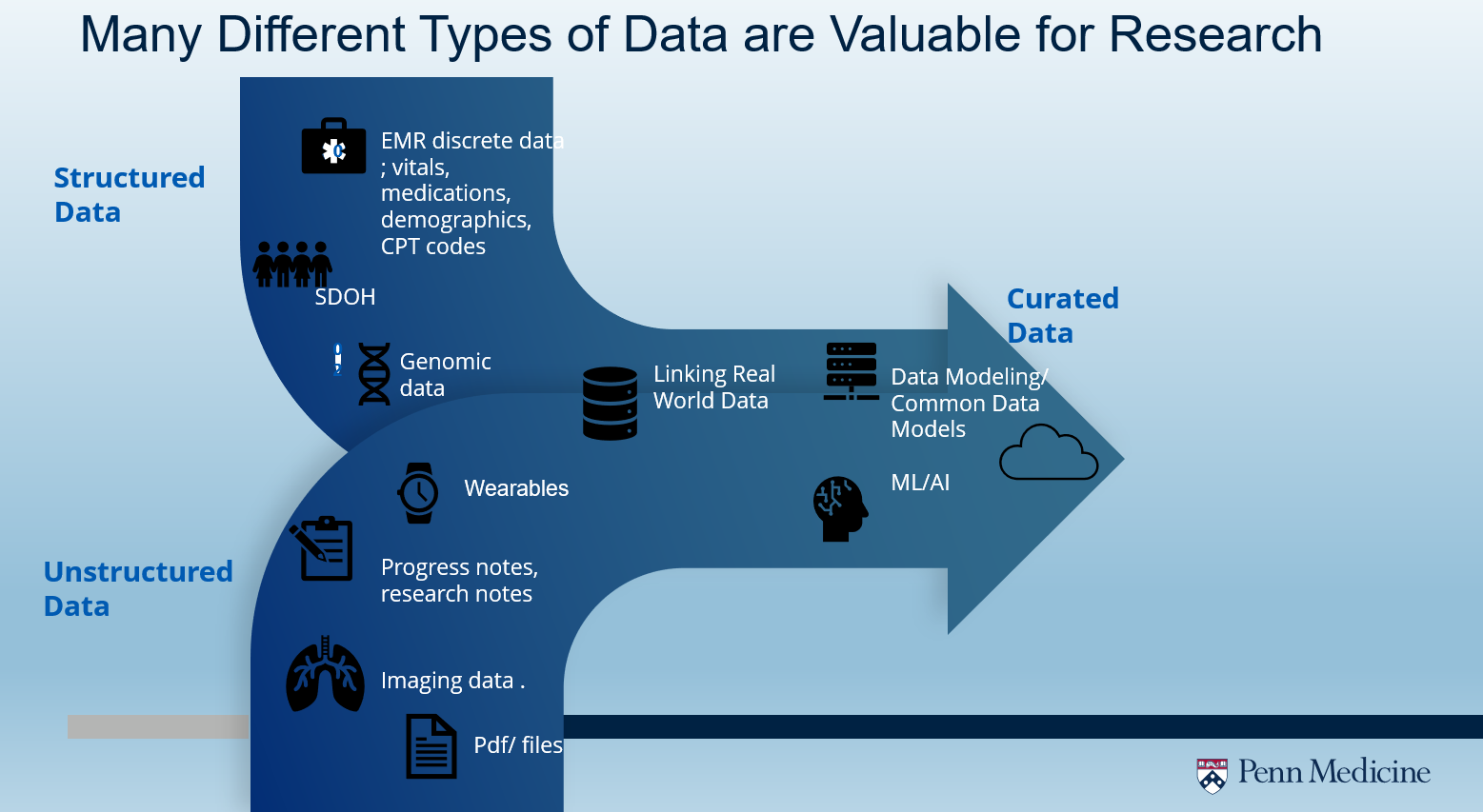
Data Types and Access
Penn Medicine has many different types of patient data that are valuable for research activities.
For example:
- Structured data in the EMR such as vitals; demographics, lab values, etc.
- Unstructured data like notes, scales and questionnaires. .
- Imaging data such as patient X-rays and scans
- Curated data such as cloud-based common data models
Here at Penn Medicine, we use data for all types of research:
- Clinical trials
- Decentralized, or remote clinical trials
- Retrospective research
- Observational or longitudinal research
- Pragmatics clinical research with real world data
- Development of algorithms or Large language models (LLMs) using Artificial Intelligence (AI)
Penn Medicine Self Service Data Options
Honest Brokers
Honest Brokers are members of a department, division or core that access and broker data to research team under research IRB specifications and privacy restrictions.
A Research Honest Broker must have normal access (as part of their job responsibilities) to the data, records, specimens, etc. that they will be obtaining and de-identifying. A Research Honest Broker is an individual, acting on behalf of a researcher in a core, center, or department to collect and provide de-identified/ limited data set or identifiable clinical data /samples to the research team. In most cases, the Research Honest Broker systems are set up to obtain and provide clinical/medical records, data and specimens.
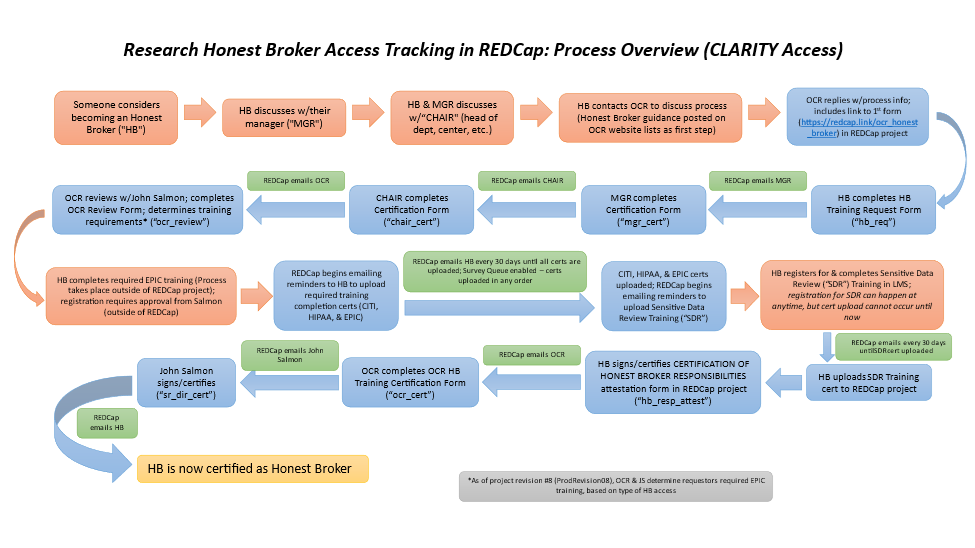
An overview of the process for access is depicted below. If interested, reach out to OCR Operations (psom-ocrops@pobox.upenn.edu)
Vendor Data Assets

TriNetX
Penn Medicine Researchers who need to access EMR data, and data beyond what is contained in the EMR, in a prep for research capacity can use a tool called TriNetX. There are several different TriNetX Networks available but two we will focus on for research are the Penn Medicine Network and the Research Network.
The Research Network has a limited data set of Penn EMR data, plus historic Penn Data Store data, mapped using The Observational Medical Outcomes Partnership (OMOP), Common Data Model, which is combined with EMR, and other real-world data like mortality data and claims data, from over 170 other health care organizations. The dataset is considered deidentified via an expert statistical determination. The data can be used for real world data analysis and already has some privacy protection built in. It provides a rich dataset that is not limited to just Penn data.
The Penn Medicine TriNetX Network is designed as above (limited data set of Penn EMR data, plus historic Penn Data Store data, mapped using The Observational Medical Outcomes Partnership (OMOP), Common Data Model), but only includes Penn Medicine patients. Queries can be run on this network to explore a Penn Medicine patient cohort identification question. Queries from this network can be rerun, once IRB approval is in place, to identify a research cohort to contact for recruitment and consent.
More about access, training, guidance for TriNetX.
Cosmos
Cosmos is a tool that is made up of structured patient data from participating Epic health care systems, including UPHS. It includes a limited data set of Penn Medicine data, with no direct identifiers, but does include dates and zip codes, combined with other Epic sites’ data. This is best used for cohort exploration, analytics, longitudinal studies and observational studies.
TriNetX and Cosmos tools may at face value seem similar but by exploring the toolkit more you can learn more about some of the nuances that make them both similar and different. TriNetX might be a better tool in a prep for research since there is a Penn Network and Penn patients can be isolated. Cosmos is most likely better for real world longitudinal research that focuses on just EMR/ Epic data.
More about access and training for Cosmos.
Here are some key differences that researchers should take into consideration when thinking about which tool to use. Cosmos and TriNetX may seem very similar, but there are key differences.
Refer to the table below, before deciding which tool to use:
Commonalities |
Differences |
|
‣ Allow trial matching and cohort identification for clinical trials. Provides way for clinical research sponsors to contact Penn as a potential site. ‣ Ability to query real world health care data ‣ Both support the following: hypothesis generations, QI, observational, longitudinal and retrospective research ‣ Both require researchers agree to not reidentify patients in the dataset |
‣ TriNetX allows downloads of data where the Cosmos data is confined to a set environment ‣ TriNetX is beyond US data and Cosmos is not. BecauseTriNEtX is deidentified there are European data as well as data from Asia and South America ‣ Cosmos deduplicates patients based on CareEverywhere ID which means patients in the data set are unique and not counted multiple times. Since TriNetX is deidentified, patients are not deduplicated so patients may be double counted in a data set. ‣ TriNetX data is enriched with claims and mortality data in some networks. Cosmos is only Epic EMR data. ‣ TriNetX uses Common Data models like OMOP so the data is standardized to a broad based standard. Cosmos uses and Epic specific standard. ‣ Cosmos is limited data set and includes dates and zip codes. TriNetX is deidentified. ‣ TriNetX is a lower barrier to entry since is a local account log in but Cosmos requires you have Epic EMR access, then conduct Epic based training to get access. |
IMRD- IQVIA Medical Records Database
All of Us Data
NIH developed dataset that registered researchers can access for data from surveys, genomic analysis, EHR data, physical measurements, and wearables to look at a full set of factors that influence overall health and disease. All of Us Research
How to get access to All of Us
Important notes:
- The identity verification step might fail. In this case you will need an ERACommons account.
- The training(s) will take you several hours to complete.
- There are two tiers of training, one for the EHR/survey data and one for the individual-level genomic data.
- More information is available in the All Of Us Tutorial
Some of these tools require teams to go through Data Governance review.
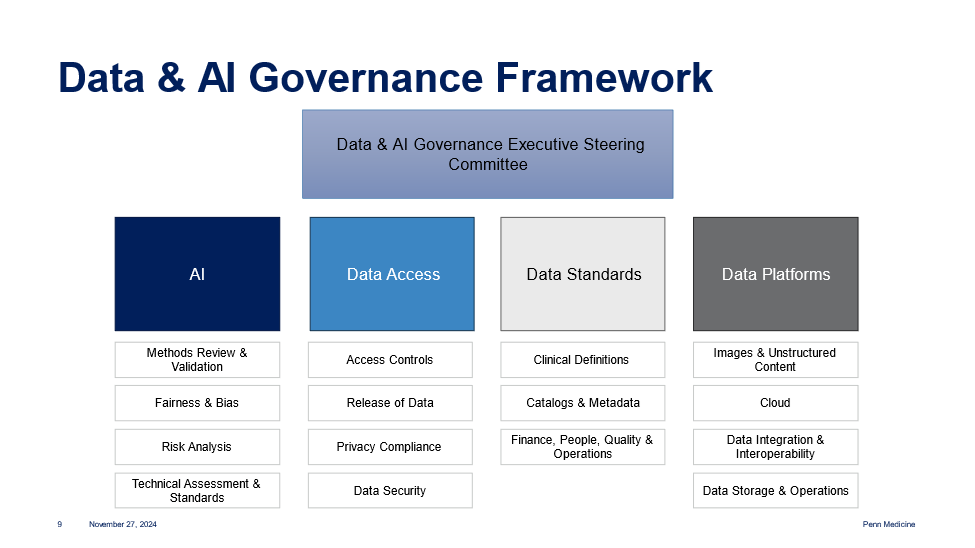
Data Access and Governance Overview