Rotation Projects
The current research focuses on the role of non-coding RNAs (microRNAs, small nuclear RNAs, etc.) and mRNA splicing in cancer pathogenesis and therapeutic resistance, with strong emphasis on chemo- and immunotherapy
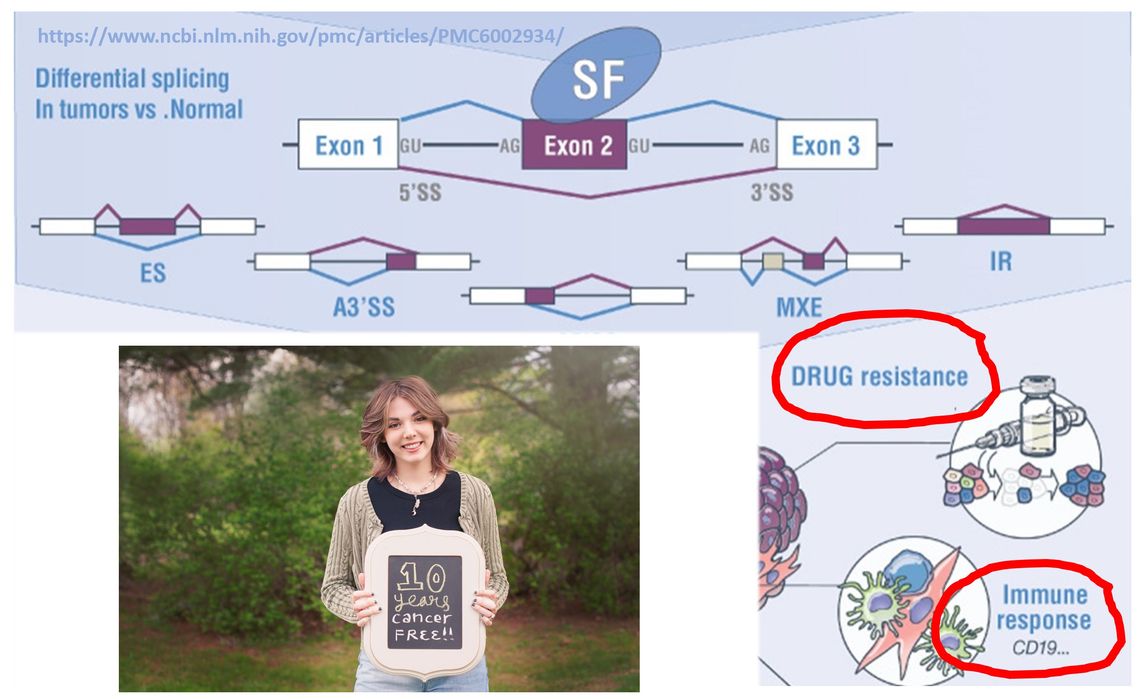
1. To investigate the effects of aberrant splicing of cell surface antigens on cancer immunotherapy (CAR T cells, antibody-drug conjugates, etc)
Suggested reading:
Sehgal P, Naqvi AS, Higgins M, Liu J, Harvey K, Jarroux J, Kim T, Mankaliye B, Mishra P, Watterson G, Fine J, Davis J, Hayer KE, Castro A, Mogbo A, Drummer C 4th, Martinez D, Koptyra MP, Ang Z, Wang K, Farrel A, Quesnel-Vallieres M, Barash Y, Spangler YB, Rokita JL, Resnick AC, Tilgner H, De Raedt T, Powell DJ Jr, Thomas-Tikhonenko A. NRCAM variant defined by microexon skipping is a targetable cell surface proteoform in high-grade gliomas. Cell Rep. 2025 Aug 7;44(8):116099
Ang Z, Paruzzo L, Hayer KE, Schmidt C, Torres Diz M, Xu F, Zankharia U, Zhang Y, Soldan S, Zheng S, Falkenstein CD, Loftus JP, Yang SY, Asnani M, King Sainos P, Pillai V, Chong E, Li MM, Tasian SK, Barash Y, Lieberman PM, Ruella M, Schuster SJ, Thomas-Tikhonenko A. Alternative splicing of its 5'-UTR limits CD20 mRNA translation and enables resistance to CD20-directed immunotherapies. Blood. 2023 Nov 16;142(20):1724-1739
Cortés-López M, Schulz L, Enculescu M, Paret C, Spiekermann B, Quesnel-Vallières M, Torres-Diz M, Unic S, Busch A, Orekhova A, Kuban M, Mesitov M, Mulorz MM, Shraim R, Kielisch F, Faber J, Barash Y, Thomas-Tikhonenko A, Zarnack K, Legewie S, König J. High-throughput mutagenesis identifies mutations and RNA-binding proteins controlling CD19 splicing and CART-19 therapy resistance. Nat Commun. 2022 Sep 22;13(1):5570
Cai T, Gouble A, Black KL, Skwarska A, Naqvi AS, Taylor D, Zhao M, Yuan Q, Sugita M, Zhang Q, Galetto R, Filipe S, Cavazos A, Han L, Kuruvilla V, Ma H, Weng C, Liu CG, Liu X, Konoplev S, Gu J, Tang G, Su X, Al-Atrash G, Ciurea S, Neelapu SS, Lane AA, Kantarjian H, Guzman ML, Pemmaraju N, Smith J, Thomas-Tikhonenko A, Konopleva M. Targeting CD123 in blastic plasmacytoid dendritic cell neoplasm using allogeneic anti-CD123 CAR T cells. Nat Commun. 2022 Apr 28;13(1):2228
Zheng S, Gillespie E, Naqvi AS, Hayer KE, Ang Z, Torres-Diz M, Quesnel-Vallières M, Hottman DA, Bagashev A, Chukinas J, Schmidt C, Asnani M, Shraim R, Taylor DM, Rheingold SR, O'Brien MM, Singh N, Lynch KW, Ruella M, Barash Y, Tasian SK, Thomas-Tikhonenko A. Modulation of CD22 Protein Expression in Childhood Leukemia by Pervasive Splicing Aberrations: Implications for CD22-Directed Immunotherapies. Blood Cancer Discov. 2022 Mar 1;3(2):103-115
Asnani M, Hayer, KE, Naqvi AS, Zheng S, Yang SY, Oldridge D, Ibrahim F, Maragkakis M, Gazzara MR, Black KL, Bagashev A, Taylor D, Mourelatos Z, Grupp SA, Barrett D, Maris JM, Sotillo E, Barash Y, Thomas-Tikhonenko, A. Retention of CD19 intron 2 contributes to CART-19 resistance in leukemias with subclonal frameshift mutations in CD19. Leukemia 2020, 34(4):1202–1207
Bagashev A, Sotillo E, Tang CA, Black KL, Perazzelli J, Seeholzer SH, Argon Y, Barrett DM, Grupp SA, Hu CA, Thomas-Tikhonenko A. CD19 alterations emerging after CD19-directed immunotherapy cause retention of the misfolded protein in the endoplasmic reticulum. Mol Cell Biol. 2018 Nov; 38(21): e00383-18
Sotillo E, Barrett DM, Black KL, Bagashev A, Oldridge D, Wu G, Sussman R, Lanauze C, Ruella M, Gazzara MR, Martinez NM, Harrington CT, Chung EY, Perazzelli J, Hofmann TJ, Maude SL, Raman P, Barrera A, Gill S, Lacey SF, Melenhorst JJ, Allman D, Jacoby E, Fry T, Mackall C, Barash Y, Lynch KW, Maris JM, Grupp SA, Thomas-Tikhonenko A. Convergence of Acquired Mutations and Alternative Splicing of CD19 Enables Resistance to CART-19 Immunotherapy. Cancer Discov. 2015 Dec;5(12):1282-95.
2. To elucidate post-transcriptional mechanisms of cancer chemoresistance (aberrant splicing, protein degradation, etc.)
Suggested reading:
Torres-Diz M, Reglero C, Falkenstein CD, Castro A, Hayer KE, Radens CM, Quesnel-Vallières M, Ang Z, Sehgal P, Li MM, Barash Y, Tasian SK, Ferrando A, Thomas-Tikhonenko A. An Alternatively Spliced Gain-of-Function NT5C2 Isoform Contributes to Chemoresistance in Acute Lymphoblastic Leukemia. Cancer Res. 2024 Aug 2;. doi: 10.1158/0008-5472.CAN-23-3804
Yang SY, Hayer KE, Fazelinia H, Spruce LA, Asnani M, Black KL, Naqvi AS, Pillai V, Barash Y, Elenitoba-Johnson KSJ, Thomas-Tikhonenko A. FBXW7β isoform drives transcriptional activation of the proinflammatory TNF cluster in human pro-B cells. Blood Adv. 2023 Apr 11;7(7):1077-1091
Harrington CT, Sotillo E, Dang CV, Thomas-Tikhonenko A. Tilting MYC toward cancer cell death. Trends Cancer. 2021 Nov;7(11):982-994
Harrington CT, Sotillo E, Robert A, Hayer KE, Bogusz AM, Psathas J, Yu D, Taylor D, Dang CV, Klein P, Hogarty MD, Geoerger B, El-Deiry WS, Wiels J, Thomas-Tikhonenko A. Transient stabilization, rather than inhibition, of MYC amplifies extrinsic apoptosis and therapeutic responses in refractory B-cell lymphoma. Leukemia. 2019 Oct 33(10): 2429–2441
3. To identify determinants of aberrant splicing in cancer, with focus on genetic variants and RNA-binding proteins
Suggested reading:
Quesnel-Vallières M, Jewell S, Lynch KW, Thomas-Tikhonenko A, Barash Y. MAJIQlopedia: an encyclopedia of RNA splicing variations in human tissues and cancer. Nucleic Acids Res. 2024 Jan 5;52(D1):D213-D221
Jha A, Quesnel-Vallières M, Wang D, Thomas-Tikhonenko A, Lynch KW, Barash Y. Identifying common transcriptome signatures of cancer by interpreting deep learning models. Genome Biol. 2022 May 17;23(1):117
Choi PS, Thomas-Tikhonenko A. RNA-binding proteins of COSMIC importance in cancer. J Clin Invest. 2021 Sep 15;131(18)
Slaff B, Radens CM, Jewell P, Jha A, Lahens NF, Grant GR, Thomas-Tikhonenko A, Lynch KW, Barash Y. MOCCASIN: a method for correcting for known and unknown confounders in RNA splicing analysis. Nat Commun. 2021 Jun 7;12(1):3353
Schulz L, Torres-Diz M, Cortés-López M, Hayer KE, Asnani M, Tasian SK, Barash Y, Sotillo E, Zarnack K, König J, Thomas-Tikhonenko A. Direct long-read RNA sequencing identifies a subset of questionable exitrons likely arising from reverse transcription artifacts. Genome Biol. 22(1):190, 2021
Black KL, Naqvi AS, Hayer KE, Yang SY, Gillespie E, Bagashev A, Pillai V, Tasian SK, Gazzara MR, Carroll M, Taylor D, Lynch KW, Barash Y, Thomas-Tikhonenko A. Aberrant splicing in B-cell acute lymphoblastic leukemia. Nucleic Acids Res. 2018, 46(21):11357–11369

