Research
Greenberg Lab
Germline mutations to the Breast Cancer 1 (BRCA1) or Breast Cancer 2 (BRCA2) genes are the major cause of hereditary breast and ovarian cancer susceptibility. Clinical BRCA1 and BRCA2 mutations render cells deficient in error-free mechanisms of DNA repair known as homologous recombination, implicating these activities in tumor suppression and response to genotoxic therapies.
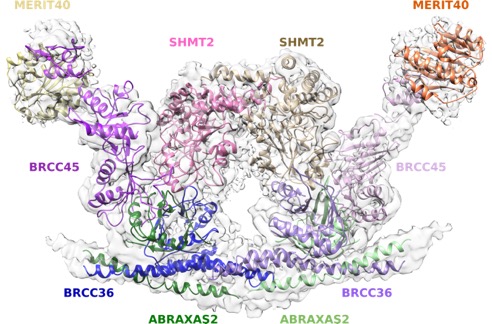
Our work has revealed a molecular understanding for how BRCA1 recognizes DNA damage and competes with opposing DNA repair proteins to control genome integrity. We have demonstrated that an interaction between the BRCA1 BRCT domain and the RAP80 ubiquitin binding protein targets BRCA1 to K63-linked ubiquitin structures present at DNA damage sites. RAP80 ubiquitin interaction motifs (UIMs) provide a ubiquitin recognition element to target BRCA1 and a K63-ubiquitin specific deubiquitinating enzyme BRCC36 to DNA double strand breaks. Each of these activities is required for appropriate DNA damage checkpoint and repair responses (Sobhian et al. Science 2007; Shao et al. Genes&Dev 2009; Tang et al. Nat Struct & Mol Biol 2013; Jiang et al. Genes Dev 2015; Zeqiraj et al. Mol Cell 2015). Cancer causing BRCA1 BRCT mutants fail to interact with RAP80 and consequently demonstrate inefficient recruitment to DNA damage sites. Moreover, germline mutations in RAP80 and Abraxas are present in familial breast cancer (Nikkila et al. Oncogene 2009; Solyom et al Sci Transl Med 2012) and biallelic BRCA1 mutations cause a new subtype of Fanconi Anemia known as Fanc-S (Domchek et al. Cancer Discov 2013; Sawyer et al Cancer Discov 2015). Thus, a series of ordered events involving ubiquitin recognition, breakdown and synthesis are required for BRCA1-dependent DNA damage responses and tumor suppression.
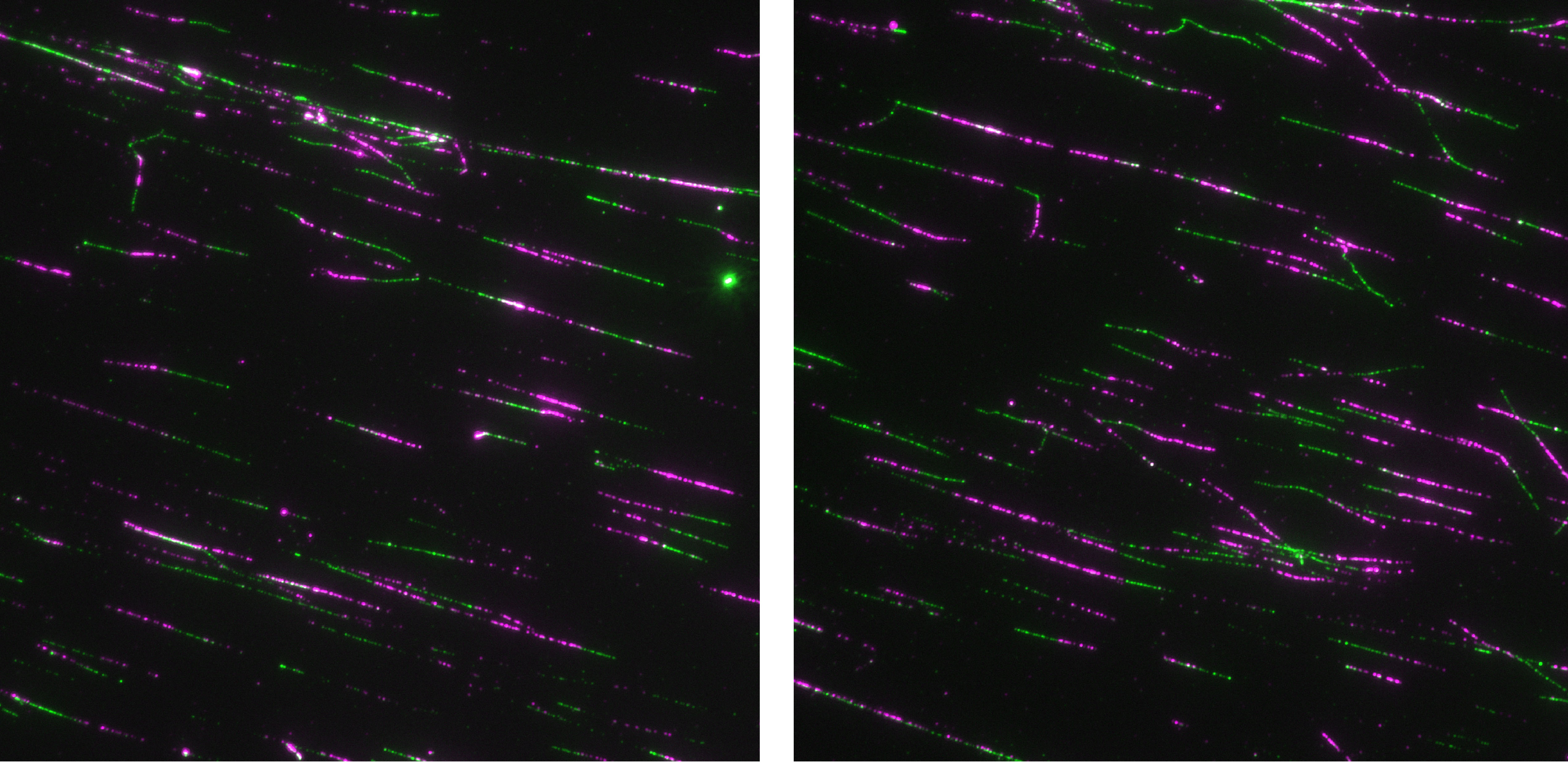
A second area of interest is the complex relationship between chromatin structure and DNA repair. We have developed several novel systems to investigate interrelationships between chromatin structure and DNA double strand break (DSB) repair. This was instrumental to our discoveries that DSBs silence transcription for multiple kilobases of chromatin in cis to the site of DNA damage (Shanbhag et al. Cell 2010), and that chromatin environment affects DNA repair mechanism choice and sensitivity to PARP inhibitors (Tang et al. Nat Struct Mol Biol 2013). More recently, we have developed methodologies to directly monitor homologous recombination at telomeres, a first for any genomic location in mammalian cells. This enabled our discovery of a novel form of homology directed repair that is responsible for alternative telomere length maintenance mechanisms in approximately 15% of human cancers (Cho et al. Cell 2014; Dilley et al. Nature 2016; Verma et al. Genes Dev 2019).
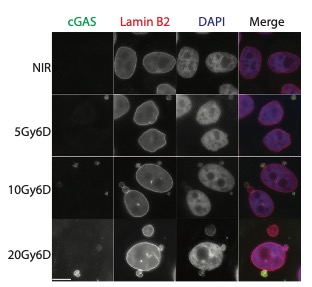
In addition to these studies involving acute DNA damage responses, we have recently determined the basis for the longstanding observation that DNA damage activates innate and adaptive immune responses (Harding et al. Nature 2017; Walden, Tian et al. Nature 2019). Our findings reveal that mitotic progression in the presence of DNA damage results in micronuclei within the cytoplasm that are recognized by the pattern recognition receptor cGAS. This produces inflammatory cytokine signals that activate anti-tumor immune responses to eradicate cells within the primary tumor and distal metastases. We will continue to use these systems to determine how DNA damage response mechanisms contributes to genome integrity, cancer etiology and response to therapy.
Black Lab
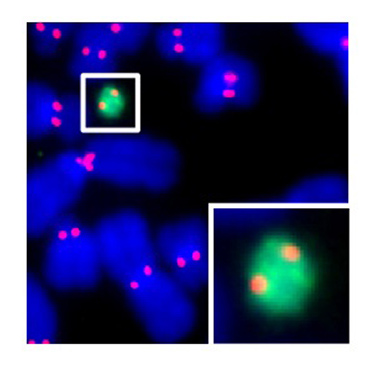
Dr. Black's laboratory is interested in how particular proteins guard the genome with emphasis on processes for detecting DNA damage and directing accurate chromosome segregation at mitosis. For the latter, the chromosomal element—the centromere—that directs this process is not defined by a particular DNA sequence. Rather, the location of the centromere is dictated by an epigenetic mark generated by one or more resident proteins. These centromeric proteins interact directly with the DNA to create a specialized chromatin compartment that is distinct from any other part of the chromosome. By taking biophysical, biochemical, and cell biological approaches, we are defining the composition and physical characteristics of the protein and protein/DNA complexes that epigenetically mark the location of the centromere on the chromosome. This work involves building centromeric chromatin from its component parts for analysis of its physical characteristics, developing biochemical assays to reconstitute steps in the process of establishing and maintaining the epigenetic mark, exploiting the power of next generation genomic and epigenomic sequencing approaches, and using animal and cell-based approaches to study the behavior of proteins involved in centromere inheritance and other essential aspects related to chromosome segregation at cell division.
Bernstein Lab
DNA double-strand breaks are the most lethal type of DNA damage. The Bernstein lab uses the budding yeast and mammalian systems to study genes whose protein products participate in accurate repair of DNA double-strand breaks, and when mutated, contribute to cancer development. More specifically, the Bernstein lab has been focusing on DNA repair genes known as RAD51 paralogs, which when mutated lead to increased breast and ovarian cancer risk. Our lab interrogates the function of these genes using genetic, biochemical, and cell biological approaches. The RAD51 paralogs function to repair DNA double-strand breaks during the high-fidelity homologous recombination pathway. At the same time, the RAD51 paralogs function to protect damaged DNA replication forks from degradation and/or enable restart of DNA replication. The RAD51 paralogs are highly conserved where they have evolved to facilitate error-free repair of DNA damage. Due to many technical challenges of studying the mammalian RAD51 paralogs, the Bernstein lab has been studying Rad51 paralog function using budding yeast. Using this system, the Bernstein lab discovered that Rad51 paralogs recognize specific DNA adducts during replication and mediate efficient replication bypass of these lesions. Since the Rad51 paralogs are conserved from yeast to humans and mutations in these proteins are observed in cancer, the Bernstein lab used yeast to determine whether mutations found in human cancers alter RAD51 paralog DNA repair functions. By screening hundreds of cancer variants in yeast, the Bernstein lab identified regions in the RAD51 paralogs that are important for their DNA repair activities in human cells. Current projects in the Bernstein lab focus on determining individuals with RAD51 paralog mutations who are at risk for cancer development and targeting these RAD51 paralog deficient tumors with new chemotherapeutic approaches. By understanding RAD51 paralog molecular function, the Bernstein lab aims to determine individuals who are at risk for cancer development and to uncover more effective targeted treatment strategies for these patients.
Brodsky Lab
The Brodsky Lab is interested in biochemical, genetic, cell biology, and systems-based approaches to investigate host-pathogen interactions. In particular, they are interested in innate immune defense against enteric bacterial pathogens. They study pattern recognition receptor and inflammatory cell death signaling in response to the gram-negative bacteria Salmonella and Yersinia, and how interactions between these innate immune responses and bacterial virulence factors influence the outcome of infection.
Busino Lab
Hematological malignancies are a group of cancers that impact the blood, bone marrow, and lymph nodes. Collectively, they comprise about 10% of all new cancer diagnoses in the United States. The ubiquitin-proteasome system (UPS) has emerged as a critical target in treating these malignancies. Drugs like thalidomide and bortezomib, which target the UPS, have shown significant improvements in patient survival. However, resistance to these therapies often develops over time. Thus, identifying new UPS enzymes as potential therapeutic targets is essential for advancing treatment options.
Our laboratory is at the forefront of research in two key areas:
- Exploring the Ubiquitin System in Hematologic Diseases: We delve into the molecular functions of ubiquitin ligases tin the context of cancer development. By integrating genetic and proteomic methods, our team is dedicated to discovering and understanding new substrate-ligase pairings. These findings are crucial in determining how they contribute to the proliferation of cancerous B-cells.
- High-Throughput Screening for Targeting Ubiquitin Ligase-Substrate Interactions: Our goal here is to develop comprehensive assays as a screening platform. These assays are designed to target specific ligase-substrate interactions. The ultimate objective of this research is to identify promising small molecules. These candidates will then undergo testing in models of leukemia and lymphoma, bringing us closer to potential new therapies.
Discher Lab
The Discher Lab is interested in topics spanning from molecular & cell biology and soft matter physics, including polymer nano-engineering. The lab's research focuses on Differentiation, Stem Cells, & Cancer: Collagens-Cytoskeleton-Nucleus-Genome connections as well as 'Marker of Self', Phagocytosis and Cell Membrane dynamics. Structures, interactions, and processes are studied in contexts ranging from short time scales (mechano-signaling, protein dynamics) to long time scales (expression, DNA repair, cell fates).
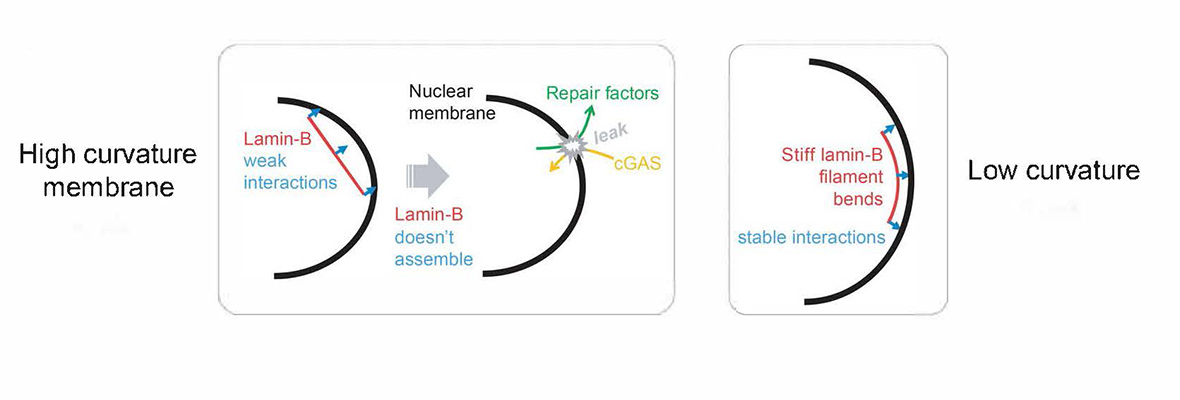
Lampson Lab
The Lampson lab pursues a variety of questions related to the cell cycle, cell division, and chromosome evolution using both mitotic and meiotic model systems. Our research is focused in the following areas:
- Mechanics of cell division, particularly interactions between chromosomes and spindle microtubules and regulation by mitotic kinases to ensure accurate chromosome segregation.
- Centromere inheritance through the germline.
- Cell biology of meiotic drive: genetic cheating in violation of Mendel's First Law.
- Optogenetic tools for cell biology.
Nader Lab
The Nader lab seeks to reconstitute the complex interface between mechanical and chemical signaling during cell migration in dense microenvironments such as interstitial spaces and solid tumors. His lab employs microfluidics, microfabrication and organ-on-a-chip devices to apply controlled and precise confinement/mechanical stress to cells and their nuclei. He combines these tools with diverse fluorescence microscopy techniques and live cell imaging to investigate how the tissue microenvironment regulates cell function through its impact on different regimes of nuclear stretching, deformation and in more extreme scenarios on nuclear envelope integrity. Our goal is to identify signaling pathways associated with nuclear mechanosensing in cells that experience different degrees of mechanical stress imposed by complex 3D microenvironments. Ultimately, this will allow us to establish a link between different degrees of nuclear deformation and cellular behaviors, from orchestrated signaling cascades to cellular perturbations and damage.
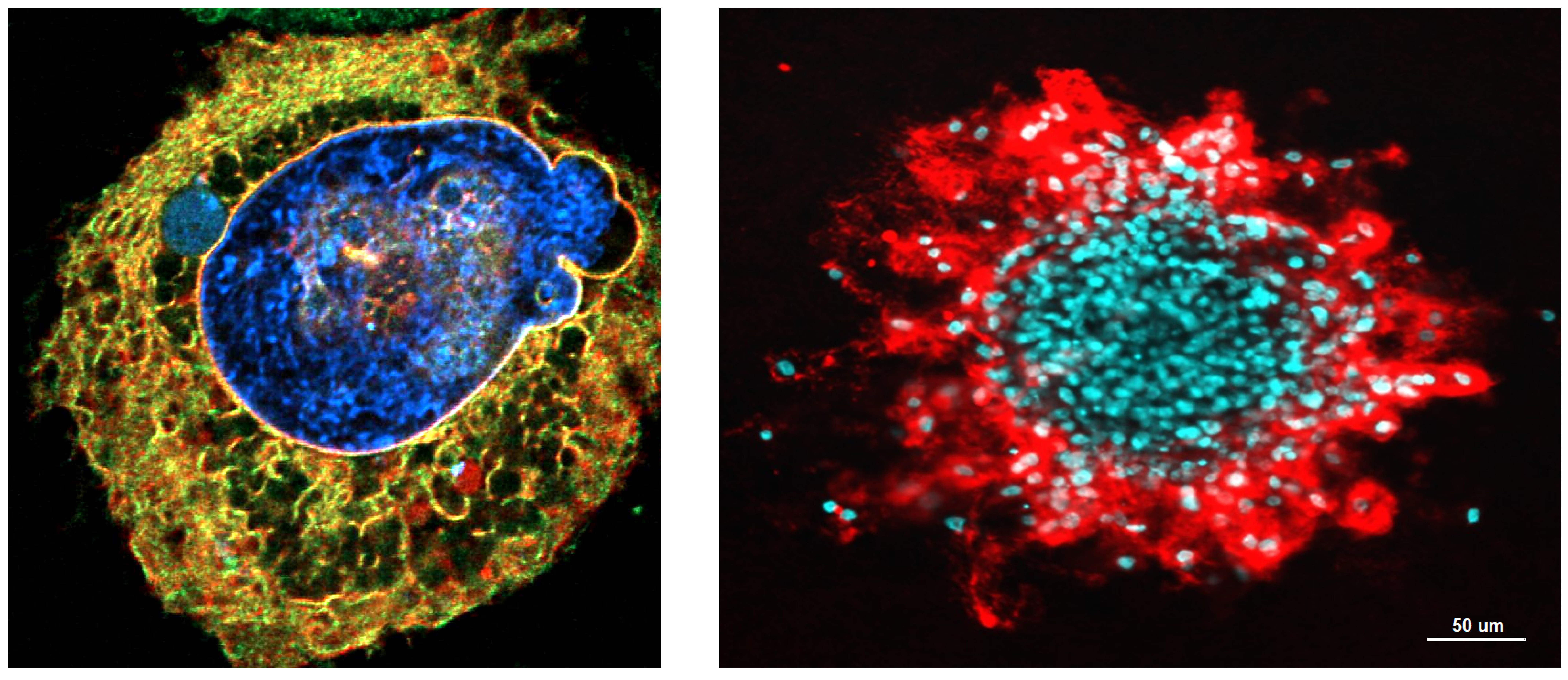
Shin Lab
The Shin Lab is interested in uncovering fundamental mechanisms of innate immunity and host defense. The lab studies how the immune system distinguishes self from non-self and tailors appropriate inflammatory responses, with a focus on pattern recognition receptors and cytosolic immune sensing pathways. One key immune pathway involves the inflammasome, a multi-protein cytosolic complex that activates the host proteases caspase-1 and caspase-11 upon cytosolic detection of bacterial products. These caspases mediate the release of IL-1 family cytokines and other inflammatory factors critical for host defense against a variety of pathogens, but overexuberant activation can lead to pathological outcomes such as septic shock. The lab is currently pursuing how mouse and human inflammasomes differentially function and how inflammasome-dependent cytokines orchestrate downstream inflammatory responses. These studies involve biochemical, cell biological, genetic, immunology, and animal-based approaches. Insight into these areas will ultimately aid in the design of more effective immune-based therapies and vaccines.
Taabazuing Lab
The Taabazuing lab utilizes molecular biology, biochemistry, chemical biology, genetics, and cell biology approaches to study the molecular regulation of cell death. Pyroptosis is an immunostimulatory type of cell death that activates the innate immune system to protect the host from invading pathogen. Pyroptosis is mediate by large multiprotein signaling complexes termed inflammasomes that activate proteases known as caspases to induce cell death. Similarly, apoptosis in the context of genotoxic stress is mediated by a large multiprotein signaling complexes termed the PIDDosome that initiates pro-survival pathways under mild genotoxic stress but initiates apoptotic cell death when genotoxic stress is severe. We are interested in understanding the intracellular triggers of these macromolecular signaling complexes, their mechanisms of assembly, and the biological professes they regulate. Understanding these processes will aid in developing therapeutics for a wide variety of human diseases, including autoinflammatory disorders and cancer.

Tong Lab
The Tong lab's research focuses on molecular hematology-oncology with an emphasis on studying signal transduction in normal blood cell development and hematological malignancies. We use genetically-engineered mouse models, bone marrow transplantation, tissue culture cells, gene transcriptional profiling, protein complex purification and mass spectrometric identification, coupled with extensive usage of molecular, cellular, and biochemical technologies.
Hundreds of billions of blood cells have to be replenished everyday. Cytokines and cytokine receptors play important roles in blood cell formation, a process known as hematopoiesis. The amplitude and duration of cytokine receptor signaling is a highly regulated process that is crucial for cytokine- governed hematopoiesis. Dysregulation of these complex signaling networks can predispose to myeloproliferative diseases and myeloid leukemia. The Tong laboratory studies kinase and ubiquitin signaling in hematopoietic stem cells, bone marrow failure, and leukemia. Hematopoietic stem cells give rise to all circulating blood cells throughout life. Maintenance of stem cell self-renewal, proliferation and differentiation is regulated by complex signaling pathways. Aberrant regulation leads to bone marrow failure or leukemia. Furthermore, genome integrity plays a critical role in the prevention of leukemia, ageing, and blood development. This postdoctoral fellow position will focus on studying the mechanisms that safeguard the genome during DNA replication and investigating how failures in these processes impact hematopoietic stem cell function that leads to bone marrow failure and/or leukemia.
Weitzman Lab
The Weitzman lab aims to understand host responses to virus infection, and the cellular environment encountered and manipulated by viruses. They study multiple viruses in an integrated experimental approach that combines biochemistry, molecular biology, genetics and cell biology. They have chosen viral models that provide tractable systems to investigate the dynamic interplay between viral genetic material and host defense strategies. Proteomic approaches are used to probe the dynamic interactions that take place on viral and cellular genomes during infection, and have uncovered ways that viruses manipulate histones and chromatin as they take control of cellular processes. The pathways illuminated are key to fighting diseases of viral infection, provide insights into fundamental processes that maintain genome instability, and have implications for the development of efficient viral vectors for gene therapy.

